The molecular signatures of compatible and incompatible pollination in Arabidopsis
- PMID: 33853522
- PMCID: PMC8048354
- DOI: 10.1186/s12864-021-07503-7
The molecular signatures of compatible and incompatible pollination in Arabidopsis
Abstract
Background: Fertilization in flowering plants depends on the early contact and acceptance of pollen grains by the receptive papilla cells of the stigma. Deciphering the specific transcriptomic response of both pollen and stigmatic cells during their interaction constitutes an important challenge to better our understanding of this cell recognition event.
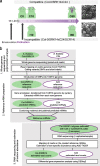
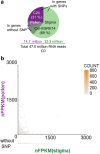
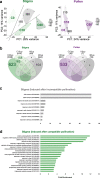
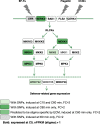
Results: Here we describe a transcriptomic analysis based on single nucleotide polymorphisms (SNPs) present in two Arabidopsis thaliana accessions, one used as female and the other as male. This strategy allowed us to distinguish 80% of transcripts according to their parental origins. We also developed a tool which predicts male/female specific expression for genes without SNP. We report an unanticipated transcriptional activity triggered in stigma upon incompatible pollination and show that following compatible interaction, components of the pattern-triggered immunity (PTI) pathway are induced on the female side.
Conclusions: Our work unveils the molecular signatures of compatible and incompatible pollinations both at the male and female side. We provide invaluable resource and tools to identify potential new molecular players involved in pollen-stigma interaction.
Keywords: Compatible / incompatible pollination; Male-female transcriptome; PTI pathway; Pollen-stigma interaction; RNA sequencing; SNPs.
Conflict of interest statement
We have no competing interests.
Figures


Similar articles
-
Transcriptional characteristics and differences in Arabidopsis stigmatic papilla cells pre- and post-pollination.Plant Cell Physiol. 2015 Apr;56(4):663-73. doi: 10.1093/pcp/pcu209. Epub 2014 Dec 19. Plant Cell Physiol. 2015. PMID: 25527828
-
Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing.Plant Cell Physiol. 2013 Nov;54(11):1894-906. doi: 10.1093/pcp/pct133. Epub 2013 Sep 20. Plant Cell Physiol. 2013. PMID: 24058146 Free PMC article.
-
Secretory activity is rapidly induced in stigmatic papillae by compatible pollen, but inhibited for self-incompatible pollen in the Brassicaceae.PLoS One. 2013 Dec 26;8(12):e84286. doi: 10.1371/journal.pone.0084286. eCollection 2013. PLoS One. 2013. PMID: 24386363 Free PMC article.
-
Exocyst, exosomes, and autophagy in the regulation of Brassicaceae pollen-stigma interactions.J Exp Bot. 2017 Dec 18;69(1):69-78. doi: 10.1093/jxb/erx340. J Exp Bot. 2017. PMID: 29036428 Review.
-
Pollen Acceptance or Rejection: A Tale of Two Pathways.Trends Plant Sci. 2016 Dec;21(12):1058-1067. doi: 10.1016/j.tplants.2016.09.004. Epub 2016 Nov 11. Trends Plant Sci. 2016. PMID: 27773670 Review.
Cited by
-
Comprehensive Analysis of BrHMPs Reveals Potential Roles in Abiotic Stress Tolerance and Pollen-Stigma Interaction in Brassica rapa.Cells. 2023 Apr 6;12(7):1096. doi: 10.3390/cells12071096. Cells. 2023. PMID: 37048168 Free PMC article.
-
Response of Fragaria vesca to projected change in temperature, water availability and concentration of CO2 in the atmosphere.Sci Rep. 2023 Jul 1;13(1):10678. doi: 10.1038/s41598-023-37901-8. Sci Rep. 2023. PMID: 37393360 Free PMC article.
-
The influence of the pollination compatibility type on the pistil S-RNase expression in European pear (Pyrus communis).Front Genet. 2024 Apr 9;15:1360332. doi: 10.3389/fgene.2024.1360332. eCollection 2024. Front Genet. 2024. PMID: 38655055 Free PMC article.
-
Transition to Self-compatibility Associated With Dominant S-allele in a Diploid Siberian Progenitor of Allotetraploid Arabidopsis kamchatica Revealed by Arabidopsis lyrata Genomes.Mol Biol Evol. 2023 Jul 5;40(7):msad122. doi: 10.1093/molbev/msad122. Mol Biol Evol. 2023. PMID: 37432770 Free PMC article.
References
-
- Bateman AJ. Self-incompatibility systems in angiosperms: III. Cruciferae. Heredity. 1955;9:53–68. doi: 10.1038/hdy.1955.2. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

