Type IV Collagen Variants in CKD: Performance of Computational Predictions for Identifying Pathogenic Variants
- PMID: 33851121
- PMCID: PMC8039416
- DOI: 10.1016/j.xkme.2020.12.007
Type IV Collagen Variants in CKD: Performance of Computational Predictions for Identifying Pathogenic Variants
Abstract
Rationale & objective: Pathogenic variants in type IV collagen have been reported to account for a significant proportion of chronic kidney disease. Accordingly, genetic testing is increasingly used to diagnose kidney diseases, but testing also may reveal rare missense variants that are of uncertain clinical significance. To aid in interpretation, computational prediction (called in silico) programs may be used to predict whether a variant is clinically important. We evaluate the performance of in silico programs for COL4A3/A4/A5 variants.
Study design setting & participants: Rare missense variants in COL4A3/A4/A5 were identified in disease cohorts, including a local focal segmental glomerulosclerosis (FSGS) cohort and publicly available disease databases, in which they are categorized as pathogenic or benign based on clinical criteria.
Tests compared & outcomes: All rare missense variants identified in the 4 disease cohorts were subjected to in silico predictions using 12 different programs. Comparisons between the predictions were compared with: (1) variant classification (pathogenic or benign) in the cohorts and (2) functional characterization in a randomly selected smaller number (17) of pathogenic or uncertain significance variants obtained from the local FSGS cohort.
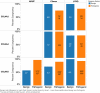
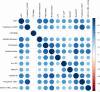
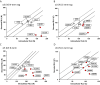
Results: In silico predictions correctly classified 75% to 97% of pathogenic and 57% to 100% of benign COL4A3/A4/A5 variants in public disease databases. The congruency of in silico predictions was similar for variants categorized as pathogenic and benign, with the exception of benign COL4A5 variants, in which disease effects were overestimated. By contrast, in silico predictions and functional characterization classified all 9 pathogenic COL4A3/A4/A5 variants correctly that were obtained from a local FSGS cohort. However, these programs also overestimated the effects of genomic variants of uncertain significance when compared with functional characterization. Each of the 12 in silico programs used yielded similar results.
Limitations: Overestimation of in silico program sensitivity given that they may have been used in the categorization of variants labeled as pathogenic in disease repositories.
Conclusions: Our results suggest that in silico predictions are sensitive but not specific to assign COL4A3/A4/A5 variant pathogenicity, with misclassification of benign variants and variants of uncertain significance. Thus, we do not recommend in silico programs but instead recommend pursuing more objective levels of evidence suggested by medical genetics guidelines.
Keywords: ARUP; Alport syndrome; ClinVar; FSGS; LOVD; computational predictions; genomics; gnomAD; in silico predictions; type IV collagen variants.
© 2021 The Authors.
Figures

Similar articles
-
Pathogenic Variants in the Genes Affected in Alport Syndrome (COL4A3-COL4A5) and Their Association With Other Kidney Conditions: A Review.Am J Kidney Dis. 2021 Dec;78(6):857-864. doi: 10.1053/j.ajkd.2021.04.017. Epub 2021 Jul 8. Am J Kidney Dis. 2021. PMID: 34245817 Review.
-
Identification of 27 Novel Variants in Genes COL4A3, COL4A4, and COL4A5 in Lithuanian Families With Alport Syndrome.Front Med (Lausanne). 2022 Mar 28;9:859521. doi: 10.3389/fmed.2022.859521. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35419377 Free PMC article.
-
X-Linked and Autosomal Recessive Alport Syndrome: Pathogenic Variant Features and Further Genotype-Phenotype Correlations.PLoS One. 2016 Sep 14;11(9):e0161802. doi: 10.1371/journal.pone.0161802. eCollection 2016. PLoS One. 2016. PMID: 27627812 Free PMC article.
-
Frequent COL4 mutations in familial microhematuria accompanied by later-onset Alport nephropathy due to focal segmental glomerulosclerosis.Clin Genet. 2017 Nov;92(5):517-527. doi: 10.1111/cge.13077. Epub 2017 Sep 25. Clin Genet. 2017. PMID: 28632965
-
Variations of type IV collagen-encoding genes in patients with histological diagnosis of focal segmental glomerulosclerosis.Pediatr Nephrol. 2020 Jun;35(6):927-936. doi: 10.1007/s00467-019-04282-y. Epub 2019 Jun 28. Pediatr Nephrol. 2020. PMID: 31254113 Review.
Cited by
-
A case report: Alport syndrome and growth hormone deficiency associated with a new COL4A4 mutation.Transl Pediatr. 2024 May 31;13(5):847-855. doi: 10.21037/tp-23-569. Epub 2024 May 17. Transl Pediatr. 2024. PMID: 38840691 Free PMC article.
-
Alport Syndrome: Clinical Spectrum and Therapeutic Advances.Kidney Med. 2023 Mar 21;5(5):100631. doi: 10.1016/j.xkme.2023.100631. eCollection 2023 May. Kidney Med. 2023. PMID: 37122389 Free PMC article. Review.
-
The Phenotypic Spectrum of COL4A3 Heterozygotes.Kidney Int Rep. 2023 Jul 25;8(10):2088-2099. doi: 10.1016/j.ekir.2023.07.010. eCollection 2023 Oct. Kidney Int Rep. 2023. PMID: 37849993 Free PMC article.
-
Interpreting ciliopathy-associated missense variants of uncertain significance (VUS) in Caenorhabditis elegans.Hum Mol Genet. 2022 May 19;31(10):1574-1587. doi: 10.1093/hmg/ddab344. Hum Mol Genet. 2022. PMID: 34964473 Free PMC article.
References
-
- Klarenbach S.W., Tonelli M., Chui B., Manns B.J. Economic evaluation of dialysis therapies. Nat Rev Nephrol. 2014;10(11):644–652. - PubMed
-
- Canadian Institute for Health Information . Canadian Institute for Health Information; Ottawa, ON: 2015. National Health Expenditure Trends, 1975 to 2015.
-
- Kashtan C.E. Alport syndrome. An inherited disorder of renal, ocular, and cochlear basement membranes. Medicine (Baltimore) 1999;78(5):338–360. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

