Coevolution underlies GPCR-G protein selectivity and functionality
- PMID: 33846507
- PMCID: PMC8041822
- DOI: 10.1038/s41598-021-87251-6
Coevolution underlies GPCR-G protein selectivity and functionality
Abstract
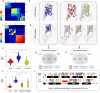
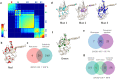
G protein-coupled receptors (GPCRs) regulate diverse physiological events, which makes them as the major targets for many approved drugs. G proteins are downstream molecules that receive signals from GPCRs and trigger cell responses. The GPCR-G protein selectivity mechanism on how they properly and timely interact is still unclear. Here, we analyzed model GPCRs (i.e. HTR, DAR) and Gα proteins with a coevolutionary tool, statistical coupling analysis. The results suggested that 5-hydroxytryptamine receptors and dopamine receptors have common conserved and coevolved residues. The Gα protein also have conserved and coevolved residues. These coevolved residues were implicated in the molecular functions of the analyzed proteins. We also found specific coevolving pairs related to the selectivity between GPCR and G protein were identified. We propose that these results would contribute to better understandings of not only the functional residues of GPCRs and Gα proteins but also GPCR-G protein selectivity mechanisms.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Sf9 cells: a versatile model system to investigate the pharmacological properties of G protein-coupled receptors.Pharmacol Ther. 2010 Dec;128(3):387-418. doi: 10.1016/j.pharmthera.2010.07.005. Epub 2010 Aug 10. Pharmacol Ther. 2010. PMID: 20705094 Review.
-
Rules and mechanisms governing G protein coupling selectivity of GPCRs.Cell Rep. 2023 Oct 31;42(10):113173. doi: 10.1016/j.celrep.2023.113173. Epub 2023 Sep 23. Cell Rep. 2023. PMID: 37742189 Free PMC article.
-
Signaling through G protein coupled receptors.Plant Signal Behav. 2009 Oct;4(10):942-7. doi: 10.4161/psb.4.10.9530. Epub 2009 Oct 14. Plant Signal Behav. 2009. PMID: 19826234 Free PMC article. Review.
-
Fine-tuning of GPCR signals by intracellular G protein modulators.Prog Mol Biol Transl Sci. 2013;115:421-53. doi: 10.1016/B978-0-12-394587-7.00010-5. Prog Mol Biol Transl Sci. 2013. PMID: 23415100 Review.
-
Regulators of G protein signaling proteins as central components of G protein-coupled receptor signaling complexes.Prog Mol Biol Transl Sci. 2009;86:49-74. doi: 10.1016/S1877-1173(09)86003-1. Epub 2009 Oct 7. Prog Mol Biol Transl Sci. 2009. PMID: 20374713 Review.
Cited by
-
DEER Analysis of GPCR Conformational Heterogeneity.Biomolecules. 2021 May 22;11(6):778. doi: 10.3390/biom11060778. Biomolecules. 2021. PMID: 34067265 Free PMC article. Review.
-
GPCRome-wide analysis of G-protein-coupling diversity using a computational biology approach.Nat Commun. 2023 Jul 19;14(1):4361. doi: 10.1038/s41467-023-40045-y. Nat Commun. 2023. PMID: 37468476 Free PMC article.
-
Evolutionary association of receptor-wide amino acids with G protein-coupling selectivity in aminergic GPCRs.Life Sci Alliance. 2022 May 25;5(10):e202201439. doi: 10.26508/lsa.202201439. Print 2022 Oct. Life Sci Alliance. 2022. PMID: 35613896 Free PMC article.
-
Evaluating conserved domains and motifs of decapod gonadotropin-releasing hormone G protein-coupled receptor superfamily.Front Endocrinol (Lausanne). 2024 Feb 20;15:1348465. doi: 10.3389/fendo.2024.1348465. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38444586 Free PMC article.
-
Hinge region mediates signal transmission of luteinizing hormone and chorionic gonadotropin receptor.Comput Struct Biotechnol J. 2022 Nov 22;20:6503-6511. doi: 10.1016/j.csbj.2022.11.039. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36467583 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

