RTKN-1/Rhotekin shields endosome-associated F-actin from disassembly to ensure endocytic recycling
- PMID: 33844824
- PMCID: PMC8047894
- DOI: 10.1083/jcb.202007149
RTKN-1/Rhotekin shields endosome-associated F-actin from disassembly to ensure endocytic recycling
Abstract
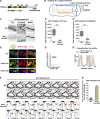
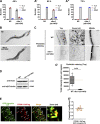
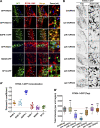
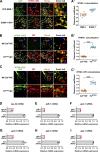
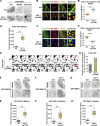
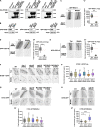
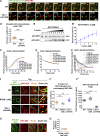
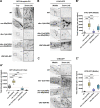
Cargo sorting and the subsequent membrane carrier formation require a properly organized endosomal actin network. To better understand the actin dynamics during endocytic recycling, we performed a genetic screen in C. elegans and identified RTKN-1/Rhotekin as a requisite to sustain endosome-associated actin integrity. Loss of RTKN-1 led to a prominent decrease in actin structures and basolateral recycling defects. Furthermore, we showed that the presence of RTKN-1 thwarts the actin disassembly competence of UNC-60A/cofilin. Consistently, in RTKN-1-deficient cells, UNC-60A knockdown replenished actin structures and alleviated the recycling defects. Notably, an intramolecular interaction within RTKN-1 could mediate the formation of oligomers. Overexpression of an RTKN-1 mutant form that lacks self-binding capacity failed to restore actin structures and recycling flow in rtkn-1 mutants. Finally, we demonstrated that SDPN-1/Syndapin acts to direct the recycling endosomal dwelling of RTKN-1 and promotes actin integrity there. Taken together, these findings consolidated the role of SDPN-1 in organizing the endosomal actin network architecture and introduced RTKN-1 as a novel regulatory protein involved in this process.
© 2021 Yan et al.
Figures





Similar articles
-
Syndapin and GTPase RAP-1 control endocytic recycling via RHO-1 and non-muscle myosin II.Curr Biol. 2023 Nov 20;33(22):4844-4856.e5. doi: 10.1016/j.cub.2023.09.051. Epub 2023 Oct 12. Curr Biol. 2023. PMID: 37832552 Free PMC article.
-
RAB-10 Promotes EHBP-1 Bridging of Filamentous Actin and Tubular Recycling Endosomes.PLoS Genet. 2016 Jun 6;12(6):e1006093. doi: 10.1371/journal.pgen.1006093. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27272733 Free PMC article.
-
WTS-1/LATS regulates endocytic recycling by restraining F-actin assembly in a synergistic manner.J Cell Sci. 2021 Dec 15;134(24):jcs259085. doi: 10.1242/jcs.259085. Epub 2021 Dec 15. J Cell Sci. 2021. PMID: 34817059
-
Endosome biogenesis is controlled by ER and the cytoskeleton at tripartite junctions.Curr Opin Cell Biol. 2023 Feb;80:102155. doi: 10.1016/j.ceb.2023.102155. Epub 2023 Feb 26. Curr Opin Cell Biol. 2023. PMID: 36848759 Review.
-
Connecting the dots: combined control of endocytic recycling and degradation.Biochem Soc Trans. 2020 Dec 18;48(6):2377-2386. doi: 10.1042/BST20180255. Biochem Soc Trans. 2020. PMID: 33300959 Free PMC article. Review.
Cited by
-
Syndapin and GTPase RAP-1 control endocytic recycling via RHO-1 and non-muscle myosin II.Curr Biol. 2023 Nov 20;33(22):4844-4856.e5. doi: 10.1016/j.cub.2023.09.051. Epub 2023 Oct 12. Curr Biol. 2023. PMID: 37832552 Free PMC article.
-
Branched-chain actin dynamics polarizes vesicle trajectories and partitions apicobasal epithelial membrane domains.Sci Adv. 2023 Jun 28;9(26):eade4022. doi: 10.1126/sciadv.ade4022. Epub 2023 Jun 28. Sci Adv. 2023. PMID: 37379384 Free PMC article.
-
Rhotekin regulates axon regeneration through the talin-Vinculin-Vinexin axis in Caenorhabditis elegans.PLoS Genet. 2023 Dec 27;19(12):e1011089. doi: 10.1371/journal.pgen.1011089. eCollection 2023 Dec. PLoS Genet. 2023. PMID: 38150455 Free PMC article.
-
PACSIN proteins in vivo: Roles in development and physiology.Acta Physiol (Oxf). 2022 Mar;234(3):e13783. doi: 10.1111/apha.13783. Epub 2022 Jan 20. Acta Physiol (Oxf). 2022. PMID: 34990060 Free PMC article. Review.
-
AP-1 Recruits SMAP-1/SMAPs to the trans-Golgi Network to Promote Sorting in Polarized Epithelia.Front Cell Dev Biol. 2021 Nov 25;9:774401. doi: 10.3389/fcell.2021.774401. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34901019 Free PMC article.
References
-
- Camera, P., da Silva J.S., Griffiths G., Giuffrida M.G., Ferrara L., Schubert V., Imarisio S., Silengo L., Dotti C.G., and Di Cunto F.. 2003. Citron-N is a neuronal Rho-associated protein involved in Golgi organization through actin cytoskeleton regulation. Nat. Cell Biol. 5:1071–1078. 10.1038/ncb1064 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

