Enzymatic bypass and the structural basis of miscoding opposite the DNA adduct 1,N2-ethenodeoxyguanosine by human DNA translesion polymerase η
- PMID: 33839151
- PMCID: PMC8121704
- DOI: 10.1016/j.jbc.2021.100642
Enzymatic bypass and the structural basis of miscoding opposite the DNA adduct 1,N2-ethenodeoxyguanosine by human DNA translesion polymerase η
Abstract
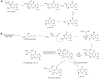
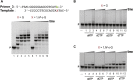
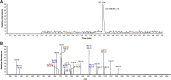
Etheno (ε)-adducts, e.g., 1,N2-ε-guanine (1,N2-ε-G) and 1,N6-ε-adenine (1,N6-ε-A), are formed through the reaction of DNA with metabolites of vinyl compounds or with lipid peroxidation products. These lesions are known to be mutagenic, but it is unknown how they lead to errors in DNA replication that are bypassed by DNA polymerases. Here we report the structural basis of misincorporation frequencies across from 1,N2-ε-G by human DNA polymerase (hpol) η. In single-nucleotide insertions opposite the adduct 1,N2-ε-G, hpol η preferentially inserted dGTP, followed by dATP, dTTP, and dCTP. This preference for purines was also seen in the first extension step. Analysis of full-length extension products by LC-MS/MS revealed that G accounted for 85% of nucleotides inserted opposite 1,N2-ε-G in single base insertion, and 63% of bases inserted in the first extension step. Extension from the correct nucleotide pair (C) was not observed, but the primer with A paired opposite 1,N2-ε-G was readily extended. Crystal structures of ternary hpol η insertion-stage complexes with nonhydrolyzable nucleotides dAMPnPP or dCMPnPP showed a syn orientation of the adduct, with the incoming A staggered between adducted base and the 5'-adjacent T, while the incoming C and adducted base were roughly coplanar. The formation of a bifurcated H-bond between incoming dAMPnPP and 1,N2-ε-G and T, compared with the single H-bond formed between incoming dCMPnPP and 1,N2-ε-G, may account for the observed facilitated insertion of dGTP and dATP. Thus, preferential insertion of purines by hpol η across from etheno adducts contributes to distinct outcomes in error-prone DNA replication.
Keywords: DNA damage; DNA polymerase; DNA replication; DNA–protein interaction; X-ray crystallography; etheno DNA adducts; mass spectrometry; translesion synthesis.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
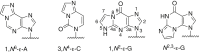


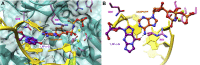

Similar articles
-
Etheno adducts: from tRNA modifications to DNA adducts and back to miscoding ribonucleotides.Genes Environ. 2021 Jun 16;43(1):24. doi: 10.1186/s41021-021-00199-x. Genes Environ. 2021. PMID: 34130743 Free PMC article. Review.
-
Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase η.J Biol Chem. 2016 Jul 1;291(27):14134-14145. doi: 10.1074/jbc.M116.732487. Epub 2016 May 16. J Biol Chem. 2016. PMID: 27226627 Free PMC article.
-
The abundant DNA adduct N7-methyl deoxyguanosine contributes to miscoding during replication by human DNA polymerase η.J Biol Chem. 2019 Jun 28;294(26):10253-10265. doi: 10.1074/jbc.RA119.008986. Epub 2019 May 17. J Biol Chem. 2019. PMID: 31101656 Free PMC article.
-
Toward understanding the mutagenicity of an environmental carcinogen: structural insights into nucleotide incorporation preferences.J Mol Biol. 2002 Sep 13;322(2):291-309. doi: 10.1016/s0022-2836(02)00751-9. J Mol Biol. 2002. PMID: 12217692
-
A rescue act: Translesion DNA synthesis past N(2) -deoxyguanosine adducts.IUBMB Life. 2015 Jul;67(7):564-74. doi: 10.1002/iub.1403. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26173005 Review.
Cited by
-
Beyond the Lesion: Back to High Fidelity DNA Synthesis.Front Mol Biosci. 2022 Jan 5;8:811540. doi: 10.3389/fmolb.2021.811540. eCollection 2021. Front Mol Biosci. 2022. PMID: 35071328 Free PMC article. Review.
-
Formation of the mutagenic DNA lesion 1,N2-ethenoguanine induced by heated cooking oil and identification of causative agents.Genes Environ. 2023 Oct 25;45(1):27. doi: 10.1186/s41021-023-00284-3. Genes Environ. 2023. PMID: 37880746 Free PMC article.
-
Etheno adducts: from tRNA modifications to DNA adducts and back to miscoding ribonucleotides.Genes Environ. 2021 Jun 16;43(1):24. doi: 10.1186/s41021-021-00199-x. Genes Environ. 2021. PMID: 34130743 Free PMC article. Review.
-
Replication Bypass of the N-(2-Deoxy-d-erythro-pentofuranosyl)-urea DNA Lesion by Human DNA Polymerase η.Biochemistry. 2024 Mar 19;63(6):754-766. doi: 10.1021/acs.biochem.3c00569. Epub 2024 Feb 27. Biochemistry. 2024. PMID: 38413007 Free PMC article.
-
Repair and tolerance of DNA damage at the replication fork: A structural perspective.Curr Opin Struct Biol. 2023 Aug;81:102618. doi: 10.1016/j.sbi.2023.102618. Epub 2023 Jun 1. Curr Opin Struct Biol. 2023. PMID: 37269798 Free PMC article. Review.
References
-
- Barbin A. Etheno-adduct-forming chemicals: From mutagenicity testing to tumor mutation spectra. Mutat. Res. 2000;462:55–69. - PubMed
-
- Singer B., Bartsch H., editors. Exocyclic DNA Adducts in Mutagenesis and Carcinogenesis, No. 150. International Agency for Research on Cancer Scientific Publications; Lyon, France: 1999.
-
- Guengerich F.P., Kim D.-H., Iwasaki M. Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem. Res. Toxicol. 1991;4:168–179. - PubMed
-
- Guengerich F.P., Persmark M., Humphreys W.G. Formation of 1,N2- and N2,3-ethenoguanine derivatives from 2-halooxiranes: Isotopic labeling studies and formation of a hemiaminal derivative of N2-(2-oxoethyl)guanine. Chem. Res. Toxicol. 1993;6:635–648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

