Surfactant cocamide monoethanolamide causes eye irritation by activating nociceptor TRPV1 channels
- PMID: 33837959
- PMCID: PMC11164132
- DOI: 10.1111/bph.15491
Surfactant cocamide monoethanolamide causes eye irritation by activating nociceptor TRPV1 channels
Abstract
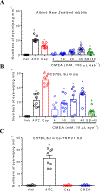
Background and purpose: Cocamide monoethanolamide (CMEA) is commonly used as a surfactant-foam booster in cosmetic formulations. Upon contact with the eye or other sensitive skin areas, CMEA elicits stinging and lasting irritation. We hypothesized a specific molecular interaction with TRPV1 channels by which CMEA caused eye irritation.
Experimental approach: Eye irritancy was evaluated using eye-wiping tests in rabbits and mice. Intracellular Ca2+ concentrations and action potentials were measured using Ca2+ imaging and current clamp respectively. Voltage clamp, site-direct mutagenesis and molecular modelling were used to identify binding pockets for CMEA on TRPV1 channels.
Key results: CMEA-induced eye irritation is ameliorated by selective ablation of TRPV1 channels.Rodents exhibit much stronger responses to CMEA than rabbits. In trigeminal ganglion neurons, CMEA induced Ca2+ influx and neuronal excitability, effects mitigated by a TRPV1 channel inhibition and absent in TRPV1 knockout neurons. In HEK-293 cells expressing TRPV1 channels, CMEA increased whole-cell currents by increasing channel open probability (EC50 = 10.2 μM), without affecting TRPV2, TRPV3, TRPV4, and TRPA1 channel activities. Lauric acid monoethanolamide (LAMEA), the most abundant constituent of CMEA, was the most efficacious and potent TRPV1 channel activator, binding to the capsaicin-binding pocket of the channel. The T550I mutants of rabbit and human TRPV1 channels exhibit much lower sensitivity to LAMEA.
Conclusions and implication: CMEA directly activates TRPV1 channels to produce eye irritation. Rabbits, the standard animal used for eye irritancy tests are poor models for evaluating human eye irritants structurally related to CMEA. Our study identifies potential alternatives to CMEA as non-irritating surfactants.
Keywords: cocamide monoethanolamide; eye irritant; transient receptor potential vanilloid type 1.
© 2021 The British Pharmacological Society.
Conflict of interest statement
Figures

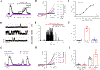


Similar articles
-
Using novel in vitro NociOcular assay based on TRPV1 channel activation for prediction of eye sting potential of baby shampoos.Toxicol Sci. 2012 Oct;129(2):325-31. doi: 10.1093/toxsci/kfs198. Epub 2012 Jun 15. Toxicol Sci. 2012. PMID: 22705807
-
Activation characteristics of transient receptor potential ankyrin 1 and its role in nociception.Am J Physiol Cell Physiol. 2011 Sep;301(3):C587-600. doi: 10.1152/ajpcell.00465.2010. Epub 2011 Jun 8. Am J Physiol Cell Physiol. 2011. PMID: 21653898 Free PMC article.
-
Identification of molecular targets for toxic action by persulfate, an industrial sulfur compound.Neurotoxicology. 2019 May;72:29-37. doi: 10.1016/j.neuro.2019.02.003. Epub 2019 Feb 6. Neurotoxicology. 2019. PMID: 30738091
-
Differential effects of TRPV channel block on polymodal activation of rat cutaneous nociceptors in vitro.Exp Brain Res. 2009 Jun;196(1):31-44. doi: 10.1007/s00221-009-1808-3. Epub 2009 Apr 30. Exp Brain Res. 2009. PMID: 19404626 Review.
-
[Activation and regulation of nociceptive transient receptor potential (TRP) channels, TRPV1 and TRPA1].Yakugaku Zasshi. 2010 Mar;130(3):289-94. doi: 10.1248/yakushi.130.289. Yakugaku Zasshi. 2010. PMID: 20190512 Review. Japanese.
References
-
- Belsito M, Hill RA, Klaassen CD, Liebler D, Marks JG Jr, Ronald C (2012). On the Safety Assessment of Ethanolamides as Used in Cosmetics. Final Amended Report
-
- Cao Z, Hulsizer S, Cui Y, Pretto DL, Kim KH, Hagerman PJ, et al. (2013). Enhanced asynchronous Ca2+ oscillations associated with impaired glutamate transport in cortical astrocytes expressing Fmr1 gene premutation expansion. Journal of Biological Chemistry 288(19): 13831–13841. 10.1074/jbc.M112.441055 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

