Biobank Scale Pharmacogenomics Informs the Genetic Underpinnings of Simvastatin Use
- PMID: 33837531
- PMCID: PMC8376807
- DOI: 10.1002/cpt.2260
Biobank Scale Pharmacogenomics Informs the Genetic Underpinnings of Simvastatin Use
Abstract
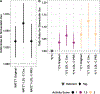
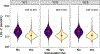
Studying drug-metabolizing enzymes, encoded by pharmacogenes, may inform biological mechanisms underlying the diseases for which a medication is prescribed. Until recently, pharmacogenes could not be studied at biobank scale. In 7,649 unrelated African-ancestry (AFR) and 326,214 unrelated European-ancestry (EUR) participants from the UK Biobank, we associated pharmacogene haplotypes from 50 genes with 265 (EUR) and 17 (AFR) medication use phenotypes using generalized linear models. In EUR, N-acetyltransferase 2 (NAT2) metabolizer phenotype and activity score were associated with simvastatin use. The dose of NAT2*1 was associated with simvastatin use when compared with NAT2*5 (the most common haplotype). This association was robust to effects of low-density lipoprotein cholesterol (LDL-C) concentration (NAT2*1 odds ratio (OR) = 1.07, 95% CI: 1.05-1.09, P = 1.14 × 10-8 ) and polygenic risk for LDL-C concentration (NAT2*1 OR = 1.09, 95% CI: 1.04-1.14, P = 2.26 × 10-4 ). Interactive effects between NAT2*1 and simvastatin use on LDL-C concentration (OR = 0.957, 95% CI: 0.916-0.998, P = 0.045) were replicated in the electronic Medical Records and Genomics Pharmacogenetic Sequencing Pilot (eMERGE-PGx) cohort (OR = 0.987, 95% CI: 0.976-0.998, P = 0.029). We used biobank-scale data to uncover and replicate an association between NAT2 locus variation and better response to statin therapy. Testing NAT2 alleles may be useful for making clinical decisions regarding the potential benefit (e.g., absolute risk reduction) in LDL-C concentration prior to statin treatment.
© 2021 The Authors. Clinical Pharmacology & Therapeutics © 2021 American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
Figures



Similar articles
-
A Mendelian Randomization Approach Using 3-HMG-Coenzyme-A Reductase Gene Variation to Evaluate the Association of Statin-Induced Low-Density Lipoprotein Cholesterol Lowering With Noncardiovascular Disease Phenotypes.JAMA Netw Open. 2021 Jun 1;4(6):e2112820. doi: 10.1001/jamanetworkopen.2021.12820. JAMA Netw Open. 2021. PMID: 34097045 Free PMC article.
-
N-acetyltransferase 1 and 2 polymorphisms in para-substituted arylamine-induced contact allergy.Br J Dermatol. 2000 Jun;142(6):1121-7. doi: 10.1046/j.1365-2133.2000.03536.x. Br J Dermatol. 2000. PMID: 10848734
-
Association between bone loss in periodontal disease and polymorphism of N-acetyltransferase (NAT2).J Clin Periodontol. 2002 Jan;29(1):21-7. doi: 10.1034/j.1600-051x.2002.290104.x. J Clin Periodontol. 2002. PMID: 11846845
-
Comparison of low-density lipoprotein cholesterol reduction after switching patients on other statins to rosuvastatin or simvastatin in a real-world clinical practice setting.Am J Manag Care. 2007 Dec;13 Suppl 10:S270-5. Am J Manag Care. 2007. PMID: 18095777 Review.
-
A meta-analysis of the association of N-acetyltransferase 2 gene (NAT2) variants with breast cancer.Am J Epidemiol. 2007 Aug 1;166(3):246-54. doi: 10.1093/aje/kwm066. Epub 2007 May 29. Am J Epidemiol. 2007. PMID: 17535831 Review.
Cited by
-
Pharmacogenomics Variability of Lipid-Lowering Therapies in Familial Hypercholesterolemia.J Pers Med. 2021 Aug 31;11(9):877. doi: 10.3390/jpm11090877. J Pers Med. 2021. PMID: 34575654 Free PMC article. Review.
-
Pharmacogenomics Informs Cardiovascular Pharmacotherapy.Methods Mol Biol. 2022;2547:201-240. doi: 10.1007/978-1-0716-2573-6_9. Methods Mol Biol. 2022. PMID: 36068466
-
From pharmacogenetics to pharmaco-omics: Milestones and future directions.HGG Adv. 2022 Mar 16;3(2):100100. doi: 10.1016/j.xhgg.2022.100100. eCollection 2022 Apr 14. HGG Adv. 2022. PMID: 35373152 Free PMC article. Review.
References
-
- Arrowsmith J & Miller P Trial watch: phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov 12, 569 (2013). - PubMed
-
- Duun Rohde PD, Sorensen P & Nyegaard M The genetic architecture of medication-use. medRxiv, 2020.09.30.20204438 (2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

