A class of independently evolved transcriptional repressors in plant RNA viruses facilitates viral infection and vector feeding
- PMID: 33836579
- PMCID: PMC7980396
- DOI: 10.1073/pnas.2016673118
A class of independently evolved transcriptional repressors in plant RNA viruses facilitates viral infection and vector feeding
Abstract
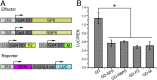
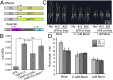
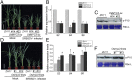
Plant viruses employ diverse virulence strategies to achieve successful infection, but there are few known general strategies of viral pathogenicity and transmission used by widely different plant viruses. Here, we report a class of independently evolved virulence factors in different plant RNA viruses which possess active transcriptional repressor activity. Rice viruses in the genera Fijivirus, Tenuivirus, and Cytorhabdovirus all have transcriptional repressors that interact in plants with the key components of jasmonic acid (JA) signaling, namely mediator subunit OsMED25, OsJAZ proteins, and OsMYC transcription factors. These transcriptional repressors can directly disassociate the OsMED25-OsMYC complex, inhibit the transcriptional activation of OsMYC, and then combine with OsJAZ proteins to cooperatively attenuate the JA pathway in a way that benefits viral infection. At the same time, these transcriptional repressors efficiently enhanced feeding by the virus insect vectors by repressing JA signaling. Our findings reveal a common strategy in unrelated plant viruses in which viral transcriptional repressors hijack and repress the JA pathway in favor of both viral pathogenicity and vector transmission.
Keywords: antiviral defense; jasmonic acid; plant viruses; transcriptional repressor; vector feeding.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures





Similar articles
-
NF-YA transcription factors suppress jasmonic acid-mediated antiviral defense and facilitate viral infection in rice.PLoS Pathog. 2022 May 13;18(5):e1010548. doi: 10.1371/journal.ppat.1010548. eCollection 2022 May. PLoS Pathog. 2022. PMID: 35560151 Free PMC article.
-
Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection.Proc Natl Acad Sci U S A. 2020 Apr 21;117(16):9112-9121. doi: 10.1073/pnas.1918254117. Epub 2020 Apr 6. Proc Natl Acad Sci U S A. 2020. PMID: 32253321 Free PMC article.
-
Exploring the shared pathogenic strategies of independently evolved effectors across distinct plant viruses.Trends Microbiol. 2024 Oct;32(10):1021-1033. doi: 10.1016/j.tim.2024.03.001. Epub 2024 Mar 22. Trends Microbiol. 2024. PMID: 38521726 Review.
-
Negative-strand RNA viruses: the plant-infecting counterparts.Virus Res. 2011 Dec;162(1-2):184-202. doi: 10.1016/j.virusres.2011.09.028. Epub 2011 Sep 22. Virus Res. 2011. PMID: 21963660 Review.
-
The Bunyavirales: The Plant-Infecting Counterparts.Viruses. 2021 May 6;13(5):842. doi: 10.3390/v13050842. Viruses. 2021. PMID: 34066457 Free PMC article. Review.
Cited by
-
NLRs derepress MED10b- and MED7-mediated repression of jasmonate-dependent transcription to activate immunity.Proc Natl Acad Sci U S A. 2023 Jul 11;120(28):e2302226120. doi: 10.1073/pnas.2302226120. Epub 2023 Jul 3. Proc Natl Acad Sci U S A. 2023. PMID: 37399403 Free PMC article.
-
Genome-Wide Analysis of the RAV Transcription Factor Genes in Rice Reveals Their Response Patterns to Hormones and Virus Infection.Viruses. 2021 Apr 25;13(5):752. doi: 10.3390/v13050752. Viruses. 2021. PMID: 33922971 Free PMC article.
-
A viral p3a protein targets and inhibits TaDOF transcription factors to promote the expression of susceptibility genes and facilitate viral infection.PLoS Pathog. 2024 Nov 7;20(11):e1012680. doi: 10.1371/journal.ppat.1012680. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39509397 Free PMC article.
-
Jasmonate Signaling Pathway Modulates Plant Defense, Growth, and Their Trade-Offs.Int J Mol Sci. 2022 Apr 1;23(7):3945. doi: 10.3390/ijms23073945. Int J Mol Sci. 2022. PMID: 35409303 Free PMC article. Review.
-
A transcription factor ZmGLK36 confers broad resistance to maize rough dwarf disease in cereal crops.Nat Plants. 2023 Oct;9(10):1720-1733. doi: 10.1038/s41477-023-01514-w. Epub 2023 Sep 14. Nat Plants. 2023. PMID: 37709955
References
-
- Wang A., Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66 (2015). - PubMed
-
- Yang Z., et al. ., Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28, 89–103.e8 (2020). - PubMed
-
- Wei T., Li Y., Rice reoviruses in insect vectors. Annu. Rev. Phytopathol. 54, 99–120 (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

