Histone deacetylases control lysine acetylation of ribosomal proteins in rice
- PMID: 33836077
- PMCID: PMC8096213
- DOI: 10.1093/nar/gkab244
Histone deacetylases control lysine acetylation of ribosomal proteins in rice
Abstract
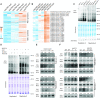
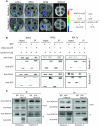
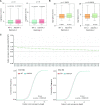
Lysine acetylation (Kac) is well known to occur in histones for chromatin function and epigenetic regulation. In addition to histones, Kac is also detected in a large number of proteins with diverse biological functions. However, Kac function and regulatory mechanism for most proteins are unclear. In this work, we studied mutation effects of rice genes encoding cytoplasm-localized histone deacetylases (HDAC) on protein acetylome and found that the HDAC protein HDA714 was a major deacetylase of the rice non-histone proteins including many ribosomal proteins (r-proteins) and translation factors that were extensively acetylated. HDA714 loss-of-function mutations increased Kac levels but reduced abundance of r-proteins. In vitro and in vivo experiments showed that HDA714 interacted with r-proteins and reduced their Kac. Substitutions of lysine by arginine (depleting Kac) in several r-proteins enhance, while mutations of lysine to glutamine (mimicking Kac) decrease their stability in transient expression system. Ribo-seq analysis revealed that the hda714 mutations resulted in increased ribosome stalling frequency. Collectively, the results uncover Kac as a functional posttranslational modification of r-proteins which is controlled by histone deacetylases, extending the role of Kac in gene expression to protein translational regulation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Global Proteome Analysis Links Lysine Acetylation to Diverse Functions in Oryza Sativa.Proteomics. 2018 Jan;18(1). doi: 10.1002/pmic.201700036. Proteomics. 2018. PMID: 29106068
-
Proteomic analysis of lysine acetylation provides strong evidence for involvement of acetylated proteins in plant meiosis and tapetum function.Plant J. 2018 Jan;93(1):142-154. doi: 10.1111/tpj.13766. Plant J. 2018. PMID: 29124795
-
A comprehensive catalog of the lysine-acetylation targets in rice (Oryza sativa) based on proteomic analyses.J Proteomics. 2016 Apr 14;138:20-9. doi: 10.1016/j.jprot.2016.01.019. Epub 2016 Feb 1. J Proteomics. 2016. PMID: 26836501
-
Advances in understanding the roles of plant HAT and HDAC in non-histone protein acetylation and deacetylation.Planta. 2024 Sep 12;260(4):93. doi: 10.1007/s00425-024-04518-8. Planta. 2024. PMID: 39264431 Review.
-
Histone Acetylation Enzymes Coordinate Metabolism and Gene Expression.Trends Plant Sci. 2015 Oct;20(10):614-621. doi: 10.1016/j.tplants.2015.07.005. Trends Plant Sci. 2015. PMID: 26440431 Review.
Cited by
-
The role of corepressor HOS15-mediated epigenetic regulation of flowering.Front Plant Sci. 2023 Jan 10;13:1101912. doi: 10.3389/fpls.2022.1101912. eCollection 2022. Front Plant Sci. 2023. PMID: 36704168 Free PMC article. Review.
-
An enhanced network of energy metabolism, lysine acetylation, and growth-promoting protein accumulation is associated with heterosis in elite hybrid rice.Plant Commun. 2023 Jul 10;4(4):100560. doi: 10.1016/j.xplc.2023.100560. Epub 2023 Feb 11. Plant Commun. 2023. PMID: 36774536 Free PMC article.
-
Ustilaginoidea virens-secreted effector Uv1809 suppresses rice immunity by enhancing OsSRT2-mediated histone deacetylation.Plant Biotechnol J. 2024 Jan;22(1):148-164. doi: 10.1111/pbi.14174. Epub 2023 Sep 16. Plant Biotechnol J. 2024. PMID: 37715970 Free PMC article.
-
Lysine acetylation of histone acetyltransferase adaptor protein ADA2 is a mechanism of metabolic control of chromatin modification in plants.Nat Plants. 2024 Mar;10(3):439-452. doi: 10.1038/s41477-024-01623-0. Epub 2024 Feb 7. Nat Plants. 2024. PMID: 38326652
-
Analysis of lysine acetylation in tomato spot wilt virus infection in Nicotiana benthamiana.Front Microbiol. 2023 Feb 2;14:1046163. doi: 10.3389/fmicb.2023.1046163. eCollection 2023. Front Microbiol. 2023. PMID: 36819054 Free PMC article.
References
-
- Verdin E., Ott M.. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015; 16:258–264. - PubMed
-
- Narita T., Weinert B.T., Choudhary C.. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019; 20:156–174. - PubMed
-
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. - PubMed
-
- Shen Y., Wei W., Zhou D.X.. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015; 20:614–621. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

