Molecular Evolution of Human Norovirus GII.2 Clusters
- PMID: 33828543
- PMCID: PMC8019798
- DOI: 10.3389/fmicb.2021.655567
Molecular Evolution of Human Norovirus GII.2 Clusters
Abstract
Background: The human norovirus GII.2 outbreak during the 2016-2017 winter season was of unprecedented scale and geographic distribution.
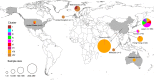
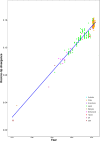
Methods: We analyzed 519 complete VP1 gene sequences of the human norovirus GII.2 genotype sampled during the 2016-2017 winter season, as well as prior (dating back to 1976) from 7 countries. Phylodynamic analyses of these sequences were performed using maximum likelihood and Bayesian statistical frameworks in order to estimate viral evolutionary and population dynamics associated with the outbreak.
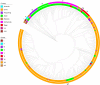
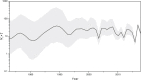
Results: Our results revealed an increase in the genetic diversity of human norovirus GII.2 during the recent Asian outbreak and diversification was characterized by at least eight distinct clusters. Bayesian estimation of viral population dynamics revealed a highly fluctuating effective population size, increasing in frequency during the past 15 years.
Conclusion: Despite an increasing viral diversity, we found no evidence of an elevated evolutionary rate or significant selection pressure in human norovirus GII.2, indicating viral evolutionary adaptation was not responsible for the volatility of or spread of the virus during this time.
Keywords: genetic diversity; human norovirus; phylogenetic analyses; positive selection; virus evolution.
Copyright © 2021 Li, Liu, Rife Magalis, Kosakovsky Pond and Volz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Genetic characterizations and molecular evolution of human norovirus GII.6 genotype during the past five decades.J Med Virol. 2023 Jun;95(6):e28876. doi: 10.1002/jmv.28876. J Med Virol. 2023. PMID: 37314035
-
Phylogenetic Analyses Suggest that Factors Other Than the Capsid Protein Play a Role in the Epidemic Potential of GII.2 Norovirus.mSphere. 2017 May 17;2(3):e00187-17. doi: 10.1128/mSphereDirect.00187-17. eCollection 2017 May-Jun. mSphere. 2017. PMID: 28529975 Free PMC article.
-
Temporal evolutionary analysis of re-emerging recombinant GII.P16_GII.2 norovirus with acute gastroenteritis in patients from Hubei Province of China, 2017.Virus Res. 2018 Apr 2;249:99-109. doi: 10.1016/j.virusres.2018.03.016. Epub 2018 Mar 28. Virus Res. 2018. PMID: 29604360
-
Temporal Evolutionary Dynamics of Norovirus GII.4 Variants in China between 2004 and 2015.PLoS One. 2016 Sep 20;11(9):e0163166. doi: 10.1371/journal.pone.0163166. eCollection 2016. PLoS One. 2016. PMID: 27649572 Free PMC article.
-
Molecular evolution of the capsid (VP1) region in human norovirus genogroup II genotype 3.Heliyon. 2020 May 3;6(5):e03835. doi: 10.1016/j.heliyon.2020.e03835. eCollection 2020 May. Heliyon. 2020. PMID: 32395646 Free PMC article.
Cited by
-
Experimental Adaptation of Murine Norovirus to Calcium Hydroxide.Front Microbiol. 2022 Mar 31;13:848439. doi: 10.3389/fmicb.2022.848439. eCollection 2022. Front Microbiol. 2022. PMID: 35432235 Free PMC article.
-
Molecular epidemiology and genetic diversity of norovirus among hospitalized children with acute gastroenteritis in Tianjin, China, 2018-2020.BMC Infect Dis. 2021 Jul 14;21(1):682. doi: 10.1186/s12879-021-06375-2. BMC Infect Dis. 2021. PMID: 34261441 Free PMC article.
-
Evolutionary analyses of emerging GII.2[P16] and GII.4 Sydney [P16] noroviruses.Virus Evol. 2022 Apr 4;8(1):veac030. doi: 10.1093/ve/veac030. eCollection 2022. Virus Evol. 2022. PMID: 35450165 Free PMC article.
-
Molecular Evolution of RNA-Dependent RNA Polymerase Region in Norovirus Genogroup I.Viruses. 2023 Jan 5;15(1):166. doi: 10.3390/v15010166. Viruses. 2023. PMID: 36680206 Free PMC article.
-
Genomic stratification and differential natural selection signatures among human norovirus genogroup II isolates.Arch Virol. 2022 May;167(5):1235-1245. doi: 10.1007/s00705-022-05396-9. Epub 2022 Mar 23. Arch Virol. 2022. PMID: 35322317 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

