From bedside to bench: regulation of host factors in SARS-CoV-2 infection
- PMID: 33828231
- PMCID: PMC8024942
- DOI: 10.1038/s12276-021-00595-x
From bedside to bench: regulation of host factors in SARS-CoV-2 infection
Abstract
The zoonotic coronavirus SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2), which causes COVID-19 (coronavirus disease-2019), has resulted in a pandemic. This has led to an urgent need to understand the molecular determinants of SARS-CoV-2 infection, factors associated with COVID-19 heterogeneity and severity, and therapeutic options for these patients. In this review, we discuss the role of host factors in SARS-CoV-2 infection and describe variations in host factor expression as mechanisms underlying the symptoms and severity of COVID-19. We focus on two host factors, angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), implicated in SARS-CoV-2 infection. We also discuss genetic variants associated with COVID-19 severity revealed in selected patients and based on genome-wide association studies (GWASs). Furthermore, we highlight important advances in cell and chromatin biology, such as single-cell RNA and chromatin sequencing and chromosomal conformation assays, as methods that may aid in the discovery of viral-host interactions in COVID-19. Understanding how regulation of host factor genes varies in physiological and pathological states might explain the heterogeneity observed in SARS-CoV-2 infection, help identify pathways for therapeutic development, and identify patients most likely to progress to severe COVID-19.
Conflict of interest statement
The authors declare no competing interests.
Figures
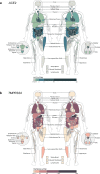
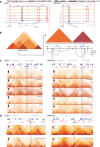
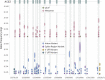

Similar articles
-
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review.
-
The contrasting role of nasopharyngeal angiotensin converting enzyme 2 (ACE2) transcription in SARS-CoV-2 infection: A cross-sectional study of people tested for COVID-19 in British Columbia, Canada.EBioMedicine. 2021 Apr;66:103316. doi: 10.1016/j.ebiom.2021.103316. Epub 2021 Apr 2. EBioMedicine. 2021. PMID: 33819740 Free PMC article.
-
ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain.J Med Virol. 2021 Feb;93(2):863-869. doi: 10.1002/jmv.26319. Epub 2020 Jul 28. J Med Virol. 2021. PMID: 32691890 Free PMC article.
-
Nasopharyngeal Expression of Angiotensin-Converting Enzyme 2 and Transmembrane Serine Protease 2 in Children within SARS-CoV-2-Infected Family Clusters.Microbiol Spectr. 2021 Dec 22;9(3):e0078321. doi: 10.1128/Spectrum.00783-21. Epub 2021 Nov 3. Microbiol Spectr. 2021. PMID: 34730438 Free PMC article.
-
Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection.J Biol Chem. 2021 Jan-Jun;296:100135. doi: 10.1074/jbc.REV120.015980. Epub 2020 Dec 6. J Biol Chem. 2021. PMID: 33268377 Free PMC article. Review.
Cited by
-
Hypothesis: Emerging Roles for Aryl Hydrocarbon Receptor in Orchestrating CoV-2-Related Inflammation.Cells. 2022 Feb 13;11(4):648. doi: 10.3390/cells11040648. Cells. 2022. PMID: 35203299 Free PMC article. Review.
-
The Relationship between COVID-19 Severity in Children and Immunoregulatory Gene Polymorphism.Viruses. 2023 Oct 14;15(10):2093. doi: 10.3390/v15102093. Viruses. 2023. PMID: 37896870 Free PMC article.
-
Appraisal of SARS-CoV-2 mutations and their impact on vaccination efficacy: an overview.J Diabetes Metab Disord. 2022 Jul 22;21(2):1763-1783. doi: 10.1007/s40200-022-01002-6. eCollection 2022 Dec. J Diabetes Metab Disord. 2022. PMID: 35891981 Free PMC article. Review.
-
Deciphering Factors Linked With Reduced Severe Acute Respiratory Syndrome Coronavirus 2 Susceptibility in the Swiss HIV Cohort Study.J Infect Dis. 2024 Aug 16;230(2):e292-e304. doi: 10.1093/infdis/jiae002. J Infect Dis. 2024. PMID: 38227786 Free PMC article.
-
Learning From Biological and Computational Machines: Importance of SARS-CoV-2 Genomic Surveillance, Mutations and Risk Stratification.Front Cell Infect Microbiol. 2021 Dec 24;11:783961. doi: 10.3389/fcimb.2021.783961. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 35047415 Free PMC article.
References
-
- Pairo-Castineira, E. et al. Genetic mechanisms of critical illness in Covid-19. Nature10.1038/s41586-020-03065-y (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

