Targeting neddylation E2s: a novel therapeutic strategy in cancer
- PMID: 33827629
- PMCID: PMC8028724
- DOI: 10.1186/s13045-021-01070-w
Targeting neddylation E2s: a novel therapeutic strategy in cancer
Abstract
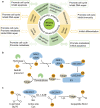
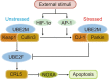
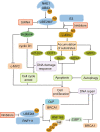
Ubiquitin-conjugating enzyme E2 M (UBE2M) and ubiquitin-conjugating enzyme E2 F (UBE2F) are the two NEDD8-conjugating enzymes of the neddylation pathway that take part in posttranslational modification and change the activity of target proteins. The activity of E2 enzymes requires both a 26-residue N-terminal docking peptide and a conserved E2 catalytic core domain, which is the basis for the transfer of neural precursor cell-expressed developmentally downregulated 8 (NEDD8). By recruiting E3 ligases and targeting cullin and non-cullin substrates, UBE2M and UBE2F play diverse biological roles. Currently, there are several inhibitors that target the UBE2M-defective in cullin neddylation protein 1 (DCN1) interaction to treat cancer. As described above, this review provides insights into the mechanism of UBE2M and UBE2F and emphasizes these two E2 enzymes as appealing therapeutic targets for the treatment of cancers.
Keywords: Anticancer treatment; Neddylation; Therapeutic targets; UBE2F; UBE2M.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Discovery of neddylation E2s inhibitors with therapeutic activity.Oncogenesis. 2023 Sep 16;12(1):45. doi: 10.1038/s41389-023-00490-2. Oncogenesis. 2023. PMID: 37717015 Free PMC article. Review.
-
Targeting DCN1-UBC12 Protein-Protein Interaction for Regulation of Neddylation Pathway.Adv Exp Med Biol. 2020;1217:349-362. doi: 10.1007/978-981-15-1025-0_20. Adv Exp Med Biol. 2020. PMID: 31898237 Review.
-
NEDD8-conjugating enzyme E2s: critical targets for cancer therapy.Cell Death Discov. 2023 Jan 23;9(1):23. doi: 10.1038/s41420-023-01337-w. Cell Death Discov. 2023. PMID: 36690633 Free PMC article. Review.
-
UBE2M Is a Stress-Inducible Dual E2 for Neddylation and Ubiquitylation that Promotes Targeted Degradation of UBE2F.Mol Cell. 2018 Jun 21;70(6):1008-1024.e6. doi: 10.1016/j.molcel.2018.06.002. Mol Cell. 2018. PMID: 29932898 Free PMC article.
-
Neddylation-CRLs regulate the functions of Treg immune cells.Bioessays. 2023 Apr;45(4):e2200222. doi: 10.1002/bies.202200222. Epub 2023 Jan 29. Bioessays. 2023. PMID: 36709423
Cited by
-
Deciphering the role of neddylation in tumor microenvironment modulation: common outcome of multiple signaling pathways.Biomark Res. 2024 Jan 8;12(1):5. doi: 10.1186/s40364-023-00545-x. Biomark Res. 2024. PMID: 38191508 Free PMC article. Review.
-
Discovery of neddylation E2s inhibitors with therapeutic activity.Oncogenesis. 2023 Sep 16;12(1):45. doi: 10.1038/s41389-023-00490-2. Oncogenesis. 2023. PMID: 37717015 Free PMC article. Review.
-
Protein neddylation and its role in health and diseases.Signal Transduct Target Ther. 2024 Apr 5;9(1):85. doi: 10.1038/s41392-024-01800-9. Signal Transduct Target Ther. 2024. PMID: 38575611 Free PMC article. Review.
-
UBE2L3 expression in human gastric cancer and its clinical significance.J Cancer Res Clin Oncol. 2024 Apr 24;150(4):210. doi: 10.1007/s00432-024-05669-7. J Cancer Res Clin Oncol. 2024. PMID: 38656363 Free PMC article.
-
Targeting cullin neddylation for cancer and fibrotic diseases.Theranostics. 2023 Sep 4;13(14):5017-5056. doi: 10.7150/thno.78876. eCollection 2023. Theranostics. 2023. PMID: 37771770 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

