C3G downregulation induces the acquisition of a mesenchymal phenotype that enhances aggressiveness of glioblastoma cells
- PMID: 33824275
- PMCID: PMC8024353
- DOI: 10.1038/s41419-021-03631-w
C3G downregulation induces the acquisition of a mesenchymal phenotype that enhances aggressiveness of glioblastoma cells
Abstract
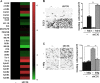
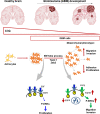
Glioblastoma (GBM) is the most aggressive tumor from the central nervous system (CNS). The current lack of efficient therapies makes essential to find new treatment strategies. C3G, a guanine nucleotide exchange factor for some Ras proteins, plays a dual role in cancer, but its function in GBM remains unknown. Database analyses revealed a reduced C3G mRNA expression in GBM patient samples. C3G protein levels were also decreased in a panel of human GBM cell lines as compared to astrocytes. Based on this, we characterized C3G function in GBM using in vitro and in vivo human GBM models. We report here that C3G downregulation promoted the acquisition of a more mesenchymal phenotype that enhanced the migratory and invasive capacity of GBM cells. This facilitates foci formation in anchorage-dependent and -independent growth assays and the generation of larger tumors in xenografts and chick chorioallantoic membrane (CAM) assays, but with a lower cell density, as proliferation was reduced. Mechanistically, C3G knock-down impairs EGFR signaling by reducing cell surface EGFR through recycling inhibition, while upregulating the activation of several other receptor tyrosine kinases (RTKs) that might promote invasion. In particular, FGF2, likely acting through FGFR1, promoted invasion of C3G-silenced GBM cells. Moreover, ERKs mediate this invasiveness, both in response to FGF2- and serum-induced chemoattraction. In conclusion, our data show the distinct dependency of GBM tumors on C3G for EGF/EGFR signaling versus other RTKs, suggesting that assessing C3G levels may discriminate GBM patient responders to different RTK inhibition protocols. Hence, patients with a low C3G expression might not respond to EGFR inhibitors.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
C3G Protein, a New Player in Glioblastoma.Int J Mol Sci. 2021 Sep 16;22(18):10018. doi: 10.3390/ijms221810018. Int J Mol Sci. 2021. PMID: 34576182 Free PMC article. Review.
-
The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases.Neoplasia. 2009 May;11(5):448-58, 2 p following 458. doi: 10.1593/neo.09230. Neoplasia. 2009. PMID: 19412429 Free PMC article.
-
C3G Is Upregulated in Hepatocarcinoma, Contributing to Tumor Growth and Progression and to HGF/MET Pathway Activation.Cancers (Basel). 2020 Aug 14;12(8):2282. doi: 10.3390/cancers12082282. Cancers (Basel). 2020. PMID: 32823931 Free PMC article.
-
Combined EGFR and autophagy modulation impairs cell migration and enhances radiosensitivity in human glioblastoma cells.J Cell Physiol. 2014 Nov;229(11):1863-73. doi: 10.1002/jcp.24640. J Cell Physiol. 2014. PMID: 24691646
-
Receptor Tyrosine Kinase Signaling and Targeting in Glioblastoma Multiforme.Int J Mol Sci. 2021 Feb 12;22(4):1831. doi: 10.3390/ijms22041831. Int J Mol Sci. 2021. PMID: 33673213 Free PMC article. Review.
Cited by
-
Characterization of prevalent tyrosine kinase inhibitors and their challenges in glioblastoma treatment.Front Chem. 2024 Jan 8;11:1325214. doi: 10.3389/fchem.2023.1325214. eCollection 2023. Front Chem. 2024. PMID: 38264122 Free PMC article. Review.
-
C3G down-regulation enhances pro-migratory and stemness properties of oval cells by promoting an epithelial-mesenchymal-like process.Int J Biol Sci. 2022 Sep 25;18(15):5873-5884. doi: 10.7150/ijbs.73192. eCollection 2022. Int J Biol Sci. 2022. PMID: 36263169 Free PMC article.
-
C3G Protein, a New Player in Glioblastoma.Int J Mol Sci. 2021 Sep 16;22(18):10018. doi: 10.3390/ijms221810018. Int J Mol Sci. 2021. PMID: 34576182 Free PMC article. Review.
-
Rab32 promotes glioblastoma migration and invasion via regulation of ERK/Drp1-mediated mitochondrial fission.Cell Death Dis. 2023 Mar 15;14(3):198. doi: 10.1038/s41419-023-05721-3. Cell Death Dis. 2023. PMID: 36922509 Free PMC article.
-
New and Old Key Players in Liver Cancer.Int J Mol Sci. 2023 Dec 5;24(24):17152. doi: 10.3390/ijms242417152. Int J Mol Sci. 2023. PMID: 38138981 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

