Phylogenetic classification of the whole-genome sequences of SARS-CoV-2 from India & evolutionary trends
- PMID: 33818474
- PMCID: PMC8184080
- DOI: 10.4103/ijmr.IJMR_3418_20
Phylogenetic classification of the whole-genome sequences of SARS-CoV-2 from India & evolutionary trends
Erratum in
-
Erratum: Phylogenetic classification of the whole-genome sequences of SARS-CoV-2 from India & evolutionary trends.Indian J Med Res. 2021 Apr;153(4):515. doi: 10.4103/0971-5916.322945. Indian J Med Res. 2021. PMID: 34380800 Free PMC article.
Abstract
Background & objectives: Several phylogenetic classification systems have been devised to trace the viral lineages of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, inconsistency in the nomenclature limits uniformity in its epidemiological understanding. This study provides an integration of existing classifications and describes evolutionary trends of the SARS-CoV-2 strains circulating in India.
Methods: The whole genomes of 330 SARS-CoV-2 samples were sequenced using next-generation sequencing (NGS). Phylogenetic and sequence analysis of a total of 3014 Indian SARS-CoV-2 sequences from 20 different States/Union Territories (January to September 2020) from the Global Initiative on Sharing All Influenza Data (GISAID) database was performed to observe the clustering of Nextstrain and Phylogenetic Assignment of Named Global Outbreak LINeages (Pangolin) lineages with the GISAID clades. The identification of mutational sites under selection pressure was performed using Mixed Effects Model of Evolution and Single-Likelihood Ancestor Counting methods available in the Datamonkey server.
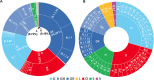
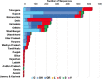
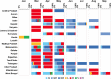
Results: Temporal data of the Indian SARS-CoV-2 genomes revealed that except for Uttarakhand, West Bengal and Haryana that showed the circulation of GISAID clade O even after July 2020, the rest of the States showed a complete switch to GR/GH clades. Pangolin lineages B.1.1.8 and B.1.113 identified within GR and GH clades, respectively, were noted to be indigenous evolutions. Sites identified to be under positive selection pressure within these clades were found to occur majorly in the non-structural proteins coded by ORF1a and ORF1b.
Interpretation & conclusions: This study interpreted the geographical and temporal dominance of SARS-CoV-2 strains in India over a period of nine months based on the GISAID classification. An integration of the GISAID, Nextstrain and Pangolin classifications is also provided. The emergence of new lineages B.1.1.8 and B.1.113 was indicative of host-specific evolution of the SARS-CoV-2 strains in India. The hotspot mutations such as those driven by positive selection need to be further characterized.
Keywords: COVID-19- nucleotide substitution; Clades; India; SARS-CoV-2; selection pressure; whole genomes.
Conflict of interest statement
None
Figures

Similar articles
-
The Novel Coronavirus Enigma: Phylogeny and Analyses of Coevolving Mutations Among the SARS-CoV-2 Viruses Circulating in India.JMIR Bioinform Biotechnol. 2020 Sep 7;1(1):e20735. doi: 10.2196/20735. eCollection 2020. JMIR Bioinform Biotechnol. 2020. PMID: 33496683 Free PMC article.
-
Evolutionary and Phylogenetic Dynamics of SARS-CoV-2 Variants: A Genetic Comparative Study of Taiyuan and Wuhan Cities of China.Viruses. 2024 Jun 3;16(6):907. doi: 10.3390/v16060907. Viruses. 2024. PMID: 38932199 Free PMC article.
-
Phylo-geo haplotype network-based characterization of SARS-CoV-2 strains circulating in India (2020-2022).Indian J Med Res. 2024 Jun;159(6):689-694. doi: 10.25259/IJMR_252_23. Indian J Med Res. 2024. PMID: 39382457 Free PMC article.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
-
COVID-19 in 16 West African Countries: An Assessment of the Epidemiology and Genetic Diversity of SARS-CoV-2 after Four Epidemic Waves.Am J Trop Med Hyg. 2023 Aug 28;109(4):861-873. doi: 10.4269/ajtmh.22-0469. Print 2023 Oct 4. Am J Trop Med Hyg. 2023. PMID: 37640294 Free PMC article. Review.
Cited by
-
Clinical Profile and Outcome of Hospitalized Confirmed Cases of Omicron Variant of SARS-CoV-2 Among Children in Pune, India.Cureus. 2022 Apr 30;14(4):e24629. doi: 10.7759/cureus.24629. eCollection 2022 Apr. Cureus. 2022. PMID: 35664398 Free PMC article.
-
Circulation and Evolution of SARS-CoV-2 in India: Let the Data Speak.Viruses. 2021 Nov 8;13(11):2238. doi: 10.3390/v13112238. Viruses. 2021. PMID: 34835044 Free PMC article.
-
Genomic epidemiology of severe acute respiratory syndrome coronavirus 2 from Theni, Tamil Nadu.J Family Med Prim Care. 2024 May;13(5):1727-1733. doi: 10.4103/jfmpc.jfmpc_1698_23. Epub 2024 May 24. J Family Med Prim Care. 2024. PMID: 38948575 Free PMC article.
-
Detection of SARS-CoV-2 variants in India from UK returnees.Infection. 2021 Dec;49(6):1355-1359. doi: 10.1007/s15010-021-01617-6. Epub 2021 Jun 23. Infection. 2021. PMID: 34160788 Free PMC article. No abstract available.
-
SARS-CoV-2 Whole-Genome Sequencing by Ion S5 Technology-Challenges, Protocol Optimization and Success Rates for Different Strains.Viruses. 2022 Jun 6;14(6):1230. doi: 10.3390/v14061230. Viruses. 2022. PMID: 35746701 Free PMC article.
References
-
- WHO Coronavirus Disease (COVID-19) Dashboard. [accessed on February 4, 2021]. Available from: https://covid19.who.int .
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

