The Novel Alpha-2 Adrenoceptor Inhibitor Beditin Reduces Cytotoxicity and Huntingtin Aggregates in Cell Models of Huntington's Disease
- PMID: 33809220
- PMCID: PMC7998230
- DOI: 10.3390/ph14030257
The Novel Alpha-2 Adrenoceptor Inhibitor Beditin Reduces Cytotoxicity and Huntingtin Aggregates in Cell Models of Huntington's Disease
Abstract
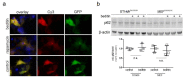
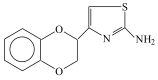
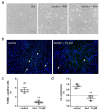
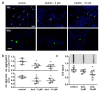
Huntington's disease (HD) is a monogenetic neurodegenerative disorder characterized by the accumulation of polyglutamine-expanded huntingtin (mHTT). There is currently no cure, and therefore disease-slowing remedies are sought to alleviate symptoms of the multifaceted disorder. Encouraging findings in Alzheimer's and Parkinson's disease on alpha-2 adrenoceptor (α2-AR) inhibition have shown neuroprotective and aggregation-reducing effects in cell and animal models. Here, we analyzed the effect of beditin, a novel α2- adrenoceptor (AR) antagonist, on cell viability and mHTT protein levels in cell models of HD using Western blot, time-resolved Foerster resonance energy transfer (TR-FRET), lactate dehydrogenase (LDH) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) cytotoxicity assays. Beditin decreases cytotoxicity, as measured by TUNEL staining and LDH release, in a neuronal progenitor cell model (STHdh cells) of HD and decreases the aggregation propensity of HTT exon 1 fragments in an overexpression model using human embryonic kidney (HEK) 293T cells. α2-AR is a promising therapeutic target for further characterization in HD models. Our data allow us to suggest beditin as a valuable candidate for the pharmaceutical manipulation of α2-AR, as it is capable of modulating neuronal cell survival and the level of mHTT.
Keywords: Huntington’s disease; alpha-2 adrenoceptor; autophagy; beditin; huntingtin; neurodegeneration; neuronal cell survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
A novel human embryonic stem cell-derived Huntington's disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration.FASEB J. 2013 May;27(5):1820-9. doi: 10.1096/fj.12-219220. Epub 2013 Jan 16. FASEB J. 2013. PMID: 23325320
-
Oleuropein enhances proteasomal activity and reduces mutant huntingtin-induced cytotoxicity.Front Pharmacol. 2024 Sep 13;15:1459909. doi: 10.3389/fphar.2024.1459909. eCollection 2024. Front Pharmacol. 2024. PMID: 39351099 Free PMC article.
-
Amelioration of Huntington's disease phenotype in astrocytes derived from iPSC-derived neural progenitor cells of Huntington's disease monkeys.PLoS One. 2019 Mar 21;14(3):e0214156. doi: 10.1371/journal.pone.0214156. eCollection 2019. PLoS One. 2019. PMID: 30897183 Free PMC article.
-
Molecular Strategies to Target Protein Aggregation in Huntington's Disease.Front Mol Biosci. 2021 Nov 12;8:769184. doi: 10.3389/fmolb.2021.769184. eCollection 2021. Front Mol Biosci. 2021. PMID: 34869596 Free PMC article. Review.
-
The P42 peptide and Peptide-based therapies for Huntington's disease.Orphanet J Rare Dis. 2016 Mar 17;11:24. doi: 10.1186/s13023-016-0405-3. Orphanet J Rare Dis. 2016. PMID: 26984770 Free PMC article. Review.
Cited by
-
How does the age of control individuals hinder the identification of target genes for Huntington's disease?Front Genet. 2024 Jun 20;15:1377237. doi: 10.3389/fgene.2024.1377237. eCollection 2024. Front Genet. 2024. PMID: 38978875 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

