Multi-Drug Featurization and Deep Learning Improve Patient-Specific Predictions of Adverse Events
- PMID: 33807714
- PMCID: PMC7967515
- DOI: 10.3390/ijerph18052600
Multi-Drug Featurization and Deep Learning Improve Patient-Specific Predictions of Adverse Events
Erratum in
-
Correction: Anastopoulos et al. Multi-Drug Featurization and Deep Learning Improve Patient-Specific Predictions of Adverse Events. Int. J. Environ. Res. Public Health 2021, 18, 2600.Int J Environ Res Public Health. 2022 Apr 1;19(7):4216. doi: 10.3390/ijerph19074216. Int J Environ Res Public Health. 2022. PMID: 35410103 Free PMC article.
Abstract
While the clinical approval process is able to filter out medications whose utility does not offset their adverse drug reaction profile in humans, it is not well suited to characterizing lower frequency issues and idiosyncratic multi-drug interactions that can happen in real world diverse patient populations. With a growing abundance of real-world evidence databases containing hundreds of thousands of patient records, it is now feasible to build machine learning models that incorporate individual patient information to provide personalized adverse event predictions. In this study, we build models that integrate patient specific demographic, clinical, and genetic features (when available) with drug structure to predict adverse drug reactions. We develop an extensible graph convolutional approach to be able to integrate molecular effects from the variable number of medications a typical patient may be taking. Our model outperforms standard machine learning methods at the tasks of predicting hospitalization and death in the UK Biobank dataset yielding an R2 of 0.37 and an AUC of 0.90, respectively. We believe our model has potential for evaluating new therapeutic compounds for individualized toxicities in real world diverse populations. It can also be used to prioritize medications when there are multiple options being considered for treatment.
Keywords: FDA FAERS; UK Biobank; adverse events; graph convolution; neural networks; real world evidence.
Conflict of interest statement
A.D. and K.D. are equity holders in Coral Genomics, Inc.
Figures


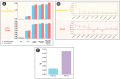
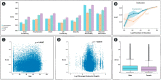
Similar articles
-
Knowledge graph prediction of unknown adverse drug reactions and validation in electronic health records.Sci Rep. 2017 Nov 27;7(1):16416. doi: 10.1038/s41598-017-16674-x. Sci Rep. 2017. PMID: 29180758 Free PMC article.
-
A study of deep learning approaches for medication and adverse drug event extraction from clinical text.J Am Med Inform Assoc. 2020 Jan 1;27(1):13-21. doi: 10.1093/jamia/ocz063. J Am Med Inform Assoc. 2020. PMID: 31135882 Free PMC article.
-
Comparing Machine Learning Algorithms for Predicting Drug-Induced Liver Injury (DILI).Mol Pharm. 2020 Jul 6;17(7):2628-2637. doi: 10.1021/acs.molpharmaceut.0c00326. Epub 2020 Jun 8. Mol Pharm. 2020. PMID: 32422053 Free PMC article.
-
Next-Generation Machine Learning for Biological Networks.Cell. 2018 Jun 14;173(7):1581-1592. doi: 10.1016/j.cell.2018.05.015. Epub 2018 Jun 7. Cell. 2018. PMID: 29887378 Review.
-
Machine-learning-based adverse drug event prediction from observational health data: A review.Drug Discov Today. 2023 Sep;28(9):103715. doi: 10.1016/j.drudis.2023.103715. Epub 2023 Jul 17. Drug Discov Today. 2023. PMID: 37467879 Review.
Cited by
-
Construction and Interpretation of Prediction Model of Teicoplanin Trough Concentration via Machine Learning.Front Med (Lausanne). 2022 Mar 8;9:808969. doi: 10.3389/fmed.2022.808969. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35360734 Free PMC article.
-
Advanced gastrointestinal stromal tumor: reliable classification of imatinib plasma trough concentration via machine learning.BMC Cancer. 2024 Feb 24;24(1):264. doi: 10.1186/s12885-024-11930-6. BMC Cancer. 2024. PMID: 38402382 Free PMC article.
-
Correction: Anastopoulos et al. Multi-Drug Featurization and Deep Learning Improve Patient-Specific Predictions of Adverse Events. Int. J. Environ. Res. Public Health 2021, 18, 2600.Int J Environ Res Public Health. 2022 Apr 1;19(7):4216. doi: 10.3390/ijerph19074216. Int J Environ Res Public Health. 2022. PMID: 35410103 Free PMC article.
-
Disease- and Drug-Related Knowledge Extraction for Health Management from Online Health Communities Based on BERT-BiGRU-ATT.Int J Environ Res Public Health. 2022 Dec 9;19(24):16590. doi: 10.3390/ijerph192416590. Int J Environ Res Public Health. 2022. PMID: 36554472 Free PMC article.
-
Strengthening clinical bacteriology laboratory diagnostics to combat sepsis and antimicrobial resistance in Benin: a train-the-trainer approach.Front Med (Lausanne). 2024 Apr 19;11:1281418. doi: 10.3389/fmed.2024.1281418. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38707185 Free PMC article.
References
-
- Ksenia J.G., Carvalhoc N.R., Chipmand K.J., Denslowe N.D., Halder M., Murphy C.A., Roelofs D., Rolaki A., Schirmer K., Watanabek K.H. Development and Application of the Adverse Outcome Pathway Framework for Understanding and Predicting Chronic Toxicity: I. Challenges and Research Needs in Ecotoxicology. Chemosphere. 2015;120:764–777. - PubMed
-
- Ngufor C., Wojtusiak J., Pathak J. A Systematic Prediction of Adverse Drug Reactions Using Pre-Clinical Drug Characteristics and Spontaneous Reports. Int. Conf. Healthc. Inform. 2015 doi: 10.1109/ICHI.2015.16. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

