Caprine MAVS Is a RIG-I Interacting Type I Interferon Inducer Downregulated by Peste des Petits Ruminants Virus Infection
- PMID: 33807534
- PMCID: PMC7998690
- DOI: 10.3390/v13030409
Caprine MAVS Is a RIG-I Interacting Type I Interferon Inducer Downregulated by Peste des Petits Ruminants Virus Infection
Abstract
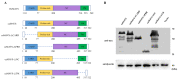
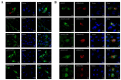
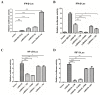
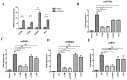
The mitochondrial antiviral-signaling protein (MAVS, also known as VISA, IPS-1, or CARDIF) plays an essential role in the type I interferon (IFN) response and in retinoic acid-inducible gene I (RIG-I) mediated antiviral innate immunity in mammals. In this study, the caprine MAVS gene (caMAVS, 1566 bp) was identified and cloned. The caMAVS shares the highest amino acid similarity (98.1%) with the predicted sheep MAVS. Confocal microscopy analysis of partial deletion mutants of caMAVS revealed that the transmembrane and the so-called Non-Characterized domains are indispensable for intracellular localization to mitochondria. Overexpression of caMAVS in caprine endometrial epithelial cells up-regulated the mRNA levels of caprine interferon-stimulated genes. We concluded that caprine MAVS mediates the activation of the type I IFN pathway. We further demonstrated that both the CARD-like domain and the transmembrane domain of caMAVS were essential for the activation of the IFN-β promotor. The interaction between caMAVS and caprine RIG-I and the vital role of the CARD and NC domain in this interaction was demonstrated by co-immunoprecipitation. Upon infection with the Peste des Petits Ruminants Virus (PPRV, genus Morbillivirus), the level of MAVS was greatly reduced. This reduction was prevented by the addition of the proteasome inhibitor MG132. Moreover, we found that viral protein V could interact and colocalize with MAVS. Together, we identified caMAVS as a RIG-I interactive protein involved in the activation of type I IFN pathways in caprine cells and as a target for PPRV immune evasion.
Keywords: Peste des Petits Ruminants Virus (PPRV); caprine; innate immunity; mitochondrial antiviral signaling protein (MAVS).
Conflict of interest statement
The authors declare that there are no conflict of interest.
Figures



Similar articles
-
Peste des petits ruminants virus non-structural C protein inhibits the induction of interferon-β by potentially interacting with MAVS and RIG-I.Virus Genes. 2021 Feb;57(1):60-71. doi: 10.1007/s11262-020-01811-y. Epub 2021 Jan 3. Virus Genes. 2021. PMID: 33389635 Free PMC article.
-
Plasminogen activator urokinase interacts with the fusion protein and antagonizes the growth of Peste des petits ruminants virus.J Virol. 2024 Apr 16;98(4):e0014624. doi: 10.1128/jvi.00146-24. Epub 2024 Mar 5. J Virol. 2024. PMID: 38440983 Free PMC article.
-
Peste des Petits Ruminants Virus Nucleocapsid Protein Inhibits Beta Interferon Production by Interacting with IRF3 To Block Its Activation.J Virol. 2019 Jul 30;93(16):e00362-19. doi: 10.1128/JVI.00362-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31167907 Free PMC article.
-
Control of peste des petits ruminants: classical and new generation vaccines.Dev Biol (Basel). 2003;114:113-9. Dev Biol (Basel). 2003. PMID: 14677682 Review.
-
Peste des Petits Ruminants Virus.Adv Virus Res. 2016;95:1-42. doi: 10.1016/bs.aivir.2016.02.001. Epub 2016 Mar 14. Adv Virus Res. 2016. PMID: 27112279 Review.
Cited by
-
Research Progress on Emerging Viral Pathogens of Small Ruminants in China during the Last Decade.Viruses. 2022 Jun 13;14(6):1288. doi: 10.3390/v14061288. Viruses. 2022. PMID: 35746759 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

