Limited Genetic Diversity Detected in Middle East Respiratory Syndrome-Related Coronavirus Variants Circulating in Dromedary Camels in Jordan
- PMID: 33807288
- PMCID: PMC8067259
- DOI: 10.3390/v13040592
Limited Genetic Diversity Detected in Middle East Respiratory Syndrome-Related Coronavirus Variants Circulating in Dromedary Camels in Jordan
Abstract
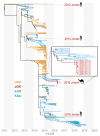
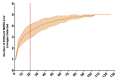
Middle East respiratory syndrome-related coronavirus (MERS-CoV) is a persistent zoonotic pathogen with frequent spillover from dromedary camels to humans in the Arabian Peninsula, resulting in limited outbreaks of MERS with a high case-fatality rate. Full genome sequence data from camel-derived MERS-CoV variants show diverse lineages circulating in domestic camels with frequent recombination. More than 90% of the available full MERS-CoV genome sequences derived from camels are from just two countries, the Kingdom of Saudi Arabia (KSA) and United Arab Emirates (UAE). In this study, we employ a novel method to amplify and sequence the partial MERS-CoV genome with high sensitivity from nasal swabs of infected camels. We recovered more than 99% of the MERS-CoV genome from field-collected samples with greater than 500 TCID50 equivalent per nasal swab from camel herds sampled in Jordan in May 2016. Our subsequent analyses of 14 camel-derived MERS-CoV genomes show a striking lack of genetic diversity circulating in Jordan camels relative to MERS-CoV genome sequences derived from large camel markets in KSA and UAE. The low genetic diversity detected in Jordan camels during our study is consistent with a lack of endemic circulation in these camel herds and reflective of data from MERS outbreaks in humans dominated by nosocomial transmission following a single introduction as reported during the 2015 MERS outbreak in South Korea. Our data suggest transmission of MERS-CoV among two camel herds in Jordan in 2016 following a single introduction event.
Keywords: MERS-CoV; coronaviruses; phylogenomics; population genomics; viral genomics; zoonoses.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Cross-sectional study of MERS-CoV-specific RNA and antibodies in animals that have had contact with MERS patients in Saudi Arabia.J Infect Public Health. 2018 May-Jun;11(3):331-338. doi: 10.1016/j.jiph.2017.09.022. Epub 2017 Oct 6. J Infect Public Health. 2018. PMID: 28993171 Free PMC article.
-
Diversity of Middle East respiratory syndrome coronaviruses in 109 dromedary camels based on full-genome sequencing, Abu Dhabi, United Arab Emirates.Emerg Microbes Infect. 2017 Nov 8;6(11):e101. doi: 10.1038/emi.2017.89. Emerg Microbes Infect. 2017. PMID: 29116217 Free PMC article.
-
Prevalence of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Abu Dhabi Emirate, United Arab Emirates.Virus Genes. 2015 Jun;50(3):509-13. doi: 10.1007/s11262-015-1174-0. Epub 2015 Feb 5. Virus Genes. 2015. PMID: 25653016 Free PMC article.
-
Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction.Pathog Glob Health. 2015;109(8):354-62. doi: 10.1080/20477724.2015.1122852. Pathog Glob Health. 2015. PMID: 26924345 Free PMC article. Review.
-
Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir.Virol J. 2016 Jun 3;13:87. doi: 10.1186/s12985-016-0544-0. Virol J. 2016. PMID: 27255185 Free PMC article. Review.
Cited by
-
Genomic surveillance of SARS-CoV-2 using long-range PCR primers.Front Microbiol. 2024 Feb 14;15:1272972. doi: 10.3389/fmicb.2024.1272972. eCollection 2024. Front Microbiol. 2024. PMID: 38440140 Free PMC article.
-
Low levels of soluble DPP4 among Saudis may have constituted a risk factor for MERS endemicity.PLoS One. 2022 Apr 12;17(4):e0266603. doi: 10.1371/journal.pone.0266603. eCollection 2022. PLoS One. 2022. PMID: 35413090 Free PMC article.
-
Special Issue "Viral Infections in Developing Countries".Viruses. 2022 Feb 16;14(2):405. doi: 10.3390/v14020405. Viruses. 2022. PMID: 35215998 Free PMC article.
-
Risk Factors for Middle East Respiratory Syndrome Coronavirus Infection among Camel Populations, Southern Jordan, 2014-2018.Emerg Infect Dis. 2021 Sep;27(9):2301-2311. doi: 10.3201/eid2709.203508. Emerg Infect Dis. 2021. PMID: 34423762 Free PMC article.
References
-
- World Health Organization . MERS Situation Update. In: Egypt C., editor. Eastern Mediterranean Regional Office. World Health Organization; Geneva, Switzerland: 2019.
-
- Yusof M.F., Queen K., Eltahir Y.M., Paden C.R., Al Hammadi Z., Tao Y., Li Y., Khalafalla A.I., Shi M., Zhang J., et al. Diversity of Middle East respiratory syndrome coronaviruses in 109 dromedary camels based on full-genome sequencing, Abu Dhabi, United Arab Emirates. Emerg. Microbes Infect. 2017;6:e101. doi: 10.1038/emi.2017.89. - DOI - PMC - PubMed
-
- van Doremalen N., Hijazeen Z.S., Holloway P., Al Omari B., McDowell C., Adney D., Talafha H.A., Guitian J., Steel J., Amarin N., et al. High Prevalence of Middle East Respiratory Coronavirus in Young Dromedary Camels in Jordan. Vector Borne Zoonotic Dis. 2017;17:155–159. doi: 10.1089/vbz.2016.2062. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

