Set-Based Rare Variant Expression Quantitative Trait Loci in Blood and Brain from Alzheimer Disease Study Participants
- PMID: 33804025
- PMCID: PMC7999141
- DOI: 10.3390/genes12030419
Set-Based Rare Variant Expression Quantitative Trait Loci in Blood and Brain from Alzheimer Disease Study Participants
Abstract
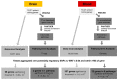
Because studies of rare variant effects on gene expression have limited power, we investigated set-based methods to identify rare expression quantitative trait loci (eQTL) related to Alzheimer disease (AD). Gene-level and pathway-level cis rare-eQTL mapping was performed genome-wide using gene expression data derived from blood donated by 713 Alzheimer's Disease Neuroimaging Initiative participants and from brain tissues donated by 475 Religious Orders Study/Memory and Aging Project participants. The association of gene or pathway expression with a set of all cis potentially regulatory low-frequency and rare variants within 1 Mb of genes was evaluated using SKAT-O. A total of 65 genes expressed in the brain were significant targets for rare expression single nucleotide polymorphisms (eSNPs) among which 17% (11/65) included established AD genes HLA-DRB1 and HLA-DRB5. In the blood, 307 genes were significant targets for rare eSNPs. In the blood and the brain, GNMT, LDHC, RBPMS2, DUS2, and HP were targets for significant eSNPs. Pathway enrichment analysis revealed significant pathways in the brain (n = 9) and blood (n = 16). Pathways for apoptosis signaling, cholecystokinin receptor (CCKR) signaling, and inflammation mediated by chemokine and cytokine signaling were common to both tissues. Significant rare eQTLs in inflammation pathways included five genes in the blood (ALOX5AP, CXCR2, FPR2, GRB2, IFNAR1) that were previously linked to AD. This study identified several significant gene- and pathway-level rare eQTLs, which further confirmed the importance of the immune system and inflammation in AD and highlighted the advantages of using a set-based eQTL approach for evaluating the effect of low-frequency and rare variants on gene expression.
Keywords: ADNI; Alzheimer disease; ROSMAP; SKAT-O; expression quantitative trait loci (eQTL); immune system; inflammation; pathways; rare variants; set-based eQTL.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
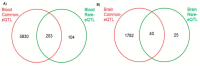
Similar articles
-
Bayesian genome-wide TWAS with reference transcriptomic data of brain and blood tissues identified 141 risk genes for Alzheimer's disease dementia.Alzheimers Res Ther. 2024 Jun 1;16(1):120. doi: 10.1186/s13195-024-01488-7. Alzheimers Res Ther. 2024. PMID: 38824563 Free PMC article.
-
CpG-related SNPs in the MS4A region have a dose-dependent effect on risk of late-onset Alzheimer disease.Aging Cell. 2019 Aug;18(4):e12964. doi: 10.1111/acel.12964. Epub 2019 May 29. Aging Cell. 2019. PMID: 31144443 Free PMC article.
-
Statistical power of transcriptome-wide association studies.Genet Epidemiol. 2022 Dec;46(8):572-588. doi: 10.1002/gepi.22491. Epub 2022 Jun 29. Genet Epidemiol. 2022. PMID: 35766062 Free PMC article.
-
Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation.PLoS One. 2015 Oct 16;10(10):e0140758. doi: 10.1371/journal.pone.0140758. eCollection 2015. PLoS One. 2015. PMID: 26474488 Free PMC article. Review.
-
Methods and Insights from Single-Cell Expression Quantitative Trait Loci.Annu Rev Genomics Hum Genet. 2023 Aug 25;24:277-303. doi: 10.1146/annurev-genom-101422-100437. Epub 2023 May 17. Annu Rev Genomics Hum Genet. 2023. PMID: 37196361 Free PMC article. Review.
Cited by
-
Identification and validation of senescence-related genes in polycystic ovary syndrome.J Ovarian Res. 2024 Jan 6;17(1):7. doi: 10.1186/s13048-023-01338-4. J Ovarian Res. 2024. PMID: 38184636 Free PMC article.
-
Editorial for the Genetics of Alzheimer's Disease Special Issue: October 2021.Genes (Basel). 2021 Nov 14;12(11):1794. doi: 10.3390/genes12111794. Genes (Basel). 2021. PMID: 34828400 Free PMC article.
-
The Alzheimer's Disease Neuroimaging Initiative in the era of Alzheimer's disease treatment: A review of ADNI studies from 2021 to 2022.Alzheimers Dement. 2024 Jan;20(1):652-694. doi: 10.1002/alz.13449. Epub 2023 Sep 12. Alzheimers Dement. 2024. PMID: 37698424 Free PMC article. Review.
-
Co-Expression Network Analysis Identifies Molecular Determinants of Loneliness Associated with Neuropsychiatric and Neurodegenerative Diseases.Int J Mol Sci. 2023 Mar 21;24(6):5909. doi: 10.3390/ijms24065909. Int J Mol Sci. 2023. PMID: 36982982 Free PMC article.
-
Innate Immune System Activation and Neuroinflammation in Down Syndrome and Neurodegeneration: Therapeutic Targets or Partners?Front Aging Neurosci. 2021 Sep 16;13:718426. doi: 10.3389/fnagi.2021.718426. eCollection 2021. Front Aging Neurosci. 2021. PMID: 34603007 Free PMC article. Review.
References
-
- Alzheimer’s Association Facts and Figures. [(accessed on 20 June 2020)];2020 Available online: https://alz.org/alzheimers-dementia/facts-figures.
-
- Sims R., van der Lee S., Naj A.C., Bellenguez C., Badarinarayan N., Jakobsdottir J., Kunkle B.W., Boland A., Raybould R., Bis J.C., et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. - DOI - PMC - PubMed
-
- Logue M.W., Schu M., Vardarajan B.N., Farrell J., Bennett D.A., Buxbaum J.D., Byrd G.S., Ertekin-Taner N., Evans D., Foroud T., et al. Two rare AKAP9 variants are associated with Alzheimer disease in African Americans. Alzheimers Dement. 2014;10:609–618. doi: 10.1016/j.jalz.2014.06.010. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

