Microscopic Interactions of Melatonin, Serotonin and Tryptophan with Zwitterionic Phospholipid Membranes
- PMID: 33799606
- PMCID: PMC8001758
- DOI: 10.3390/ijms22062842
Microscopic Interactions of Melatonin, Serotonin and Tryptophan with Zwitterionic Phospholipid Membranes
Abstract
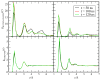
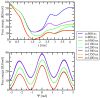
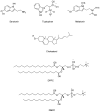
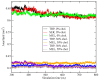
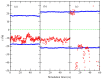
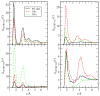
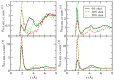
The interactions at the atomic level between small molecules and the main components of cellular plasma membranes are crucial for elucidating the mechanisms allowing for the entrance of such small species inside the cell. We have performed molecular dynamics and metadynamics simulations of tryptophan, serotonin, and melatonin at the interface of zwitterionic phospholipid bilayers. In this work, we will review recent computer simulation developments and report microscopic properties, such as the area per lipid and thickness of the membranes, atomic radial distribution functions, angular orientations, and free energy landscapes of small molecule binding to the membrane. Cholesterol affects the behaviour of the small molecules, which are mainly buried in the interfacial regions. We have observed a competition between the binding of small molecules to phospholipids and cholesterol through lipidic hydrogen-bonds. Free energy barriers that are associated to translational and orientational changes of melatonin have been found to be between 10-20 kJ/mol for distances of 1 nm between melatonin and the center of the membrane. Corresponding barriers for tryptophan and serotonin that are obtained from reversible work methods are of the order of 10 kJ/mol and reveal strong hydrogen bonding between such species and specific phospholipid sites. The diffusion of tryptophan and melatonin is of the order of 10-7 cm2/s for the cholesterol-free and cholesterol-rich setups.
Keywords: melatonin; phospholipid membrane; serotonin; tryptophan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Binding and dynamics of melatonin at the interface of phosphatidylcholine-cholesterol membranes.PLoS One. 2019 Nov 7;14(11):e0224624. doi: 10.1371/journal.pone.0224624. eCollection 2019. PLoS One. 2019. PMID: 31697738 Free PMC article.
-
Effects of cholesterol on the binding of the precursor neurotransmitter tryptophan to zwitterionic membranes.J Chem Phys. 2018 Oct 28;149(16):164906. doi: 10.1063/1.5029430. J Chem Phys. 2018. PMID: 30384712
-
The effects of melatonin, serotonin, tryptophan and NAS on the biophysical properties of DPPC monolayers.Biochim Biophys Acta Biomembr. 2020 Sep 1;1862(9):183363. doi: 10.1016/j.bbamem.2020.183363. Epub 2020 May 22. Biochim Biophys Acta Biomembr. 2020. PMID: 32450141
-
The organization of melatonin in lipid membranes.Biochim Biophys Acta. 2015 Apr;1848(4):1032-40. doi: 10.1016/j.bbamem.2015.01.006. Epub 2015 Jan 17. Biochim Biophys Acta. 2015. PMID: 25602914
-
Membrane simulations: bigger and better?Curr Opin Struct Biol. 2000 Apr;10(2):174-81. doi: 10.1016/s0959-440x(00)00066-x. Curr Opin Struct Biol. 2000. PMID: 10753807 Review.
Cited by
-
In Silico Drug Design of Benzothiadiazine Derivatives Interacting with Phospholipid Cell Membranes.Membranes (Basel). 2022 Mar 17;12(3):331. doi: 10.3390/membranes12030331. Membranes (Basel). 2022. PMID: 35323806 Free PMC article.
References
-
- Mouritsen O.G. Life-as a Matter of Fat. Springer-Verlag; Heidelberg, Germany: 2005.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

