A high-density genetic map construction and sex-related loci identification in Chinese Giant salamander
- PMID: 33794798
- PMCID: PMC8017863
- DOI: 10.1186/s12864-021-07550-0
A high-density genetic map construction and sex-related loci identification in Chinese Giant salamander
Abstract
Background: The Chinese giant salamander Andrias davidianus is an important amphibian species in China because of its increasing economic value, protection status and special evolutionary position from aquatic to terrestrial animal. Its large genome presents challenges to genetic research. Genetic linkage mapping is an important tool for genome assembly and determination of phenotype-related loci.
Results: In this study, we constructed a high-density genetic linkage map using ddRAD sequencing technology to obtain SNP genotyping data of members from an full-sib family which sex had been determined. A total of 10,896 markers were grouped and oriented into 30 linkage groups, representing 30 chromosomes of A. davidianus. The genetic length of LGs ranged from 17.61 cM (LG30) to 280.81 cM (LG1), with a mean inter-locus distance ranging from 0.11(LG3) to 0.48 cM (LG26). The total genetic map length was 2643.10 cM with an average inter-locus distance of 0.24 cM. Three sex-related loci and four sex-related markers were found on LG6 and LG23, respectively.
Conclusion: We constructed the first High-density genetic linkage map and identified three sex-related loci in the Chinese giant salamander. Current results are expected to be a useful tool for future genomic studies aiming at the marker-assisted breeding of the species.
Keywords: Chinese giant salamander; High-density linkage map; SNP; Sex-related loci mapping; ddRAD.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
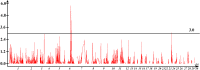
Similar articles
-
High-density genetic map construction and QTLs identification for plant height in white jute (Corchorus capsularis L.) using specific locus amplified fragment (SLAF) sequencing.BMC Genomics. 2017 May 8;18(1):355. doi: 10.1186/s12864-017-3712-8. BMC Genomics. 2017. PMID: 28482802 Free PMC article.
-
Construction of a high-density linkage map and mapping of sex determination and growth-related loci in the mandarin fish (Siniperca chuatsi).BMC Genomics. 2017 Jun 6;18(1):446. doi: 10.1186/s12864-017-3830-3. BMC Genomics. 2017. PMID: 28587594 Free PMC article.
-
Constructing a High-Density Genetic Linkage Map for Large Yellow Croaker (Larimichthys crocea) and Mapping Resistance Trait Against Ciliate Parasite Cryptocaryon irritans.Mar Biotechnol (NY). 2019 Apr;21(2):262-275. doi: 10.1007/s10126-019-09878-x. Epub 2019 Feb 19. Mar Biotechnol (NY). 2019. PMID: 30783862
-
A high-density genetic linkage map for Chinese perch (Siniperca chuatsi) using 2.3K genotyping-by-sequencing SNPs.Anim Genet. 2021 Jun;52(3):311-320. doi: 10.1111/age.13046. Epub 2021 Feb 18. Anim Genet. 2021. PMID: 33598959
-
Construction of the first high-density genetic linkage map and identification of seed yield-related QTLs and candidate genes in Elymus sibiricus, an important forage grass in Qinghai-Tibet Plateau.BMC Genomics. 2019 Nov 14;20(1):861. doi: 10.1186/s12864-019-6254-4. BMC Genomics. 2019. PMID: 31726988 Free PMC article.
Cited by
-
Construction of a high-density genetic map and QTL localization of body weight and wool production related traits in Alpine Merino sheep based on WGR.BMC Genomics. 2024 Jun 27;25(1):641. doi: 10.1186/s12864-024-10535-4. BMC Genomics. 2024. PMID: 38937677 Free PMC article.
-
Smooth Descent: A ploidy-aware algorithm to improve linkage mapping in the presence of genotyping errors.Front Genet. 2023 Mar 1;14:1049988. doi: 10.3389/fgene.2023.1049988. eCollection 2023. Front Genet. 2023. PMID: 36936433 Free PMC article.
-
Construction of a high-density genetic linkage map and QTL mapping of growth and cold tolerance traits in Takifugu fasciatus.BMC Genomics. 2023 Oct 27;24(1):645. doi: 10.1186/s12864-023-09740-4. BMC Genomics. 2023. PMID: 37891474 Free PMC article.
References
-
- Zhou ZY, Geng Y, Liu XX, Ren SY, Zhou Y, Wang KY, et al. Characterization of a ranavirus isolated from the Chinese giant salamander ( Andrias davidianus, Blanchard, 1871) in China. Aquaculture. 2013;384:66–73. doi: 10.1016/j.aquaculture.2012.12.018. - DOI
-
- Li HZ., Liu LX,, Liu XL.,Guo JS. Compared study on artificial insemination methods of Andrias davidianus. Guangdong Agric Sci,2013; 11: 128–129. [In Chinese].
-
- Xiao H., Liu J., Yang Y., Lin X. Artificial propagation of tank-cultured Chinese giant salamander Andrias davidianus. Acta Hydrobiol Sin 2006; 30: 530–534. [In Chinese].
-
- Zhang K., Wang X., Wu W., Wang ZH., Song H., et al., Advances in conservation biology of Chinese giant salamander. Biodivers Sci2002; 10: 291–297. [In Chinese].
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

