Repair of DNA Breaks by Break-Induced Replication
- PMID: 33792375
- PMCID: PMC9629446
- DOI: 10.1146/annurev-biochem-081420-095551
Repair of DNA Breaks by Break-Induced Replication
Abstract
Double-strand DNA breaks (DSBs) are the most lethal type of DNA damage, making DSB repair critical for cell survival. However, some DSB repair pathways are mutagenic and promote genome rearrangements, leading to genome destabilization. One such pathway is break-induced replication (BIR), which repairs primarily one-ended DSBs, similar to those formed by collapsed replication forks or telomere erosion. BIR is initiated by the invasion of a broken DNA end into a homologous template, synthesizes new DNA within the context of a migrating bubble, and is associated with conservative inheritance of new genetic material. This mode of synthesis is responsible for a high level of genetic instability associated with BIR. Eukaryotic BIR was initially investigated in yeast, but now it is also actively studied in mammalian systems. Additionally, a significant breakthrough has been made regarding the role of microhomology-mediated BIR in the formation of complex genomic rearrangements that underly various human pathologies.
Keywords: ALT; BIR; CRGs; DSBs; MMBIR; alternative lengthening of telomeres; break-induced replication; complex genome rearrangements; double-strand DNA breaks; microhomology-mediated break-induced replication.
Conflict of interest statement
Conflict of Interest
Authors declare no conflict of interest.
Figures

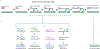

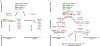
Similar articles
-
Break induced replication in eukaryotes: mechanisms, functions, and consequences.Crit Rev Biochem Mol Biol. 2017 Aug;52(4):395-413. doi: 10.1080/10409238.2017.1314444. Epub 2017 Apr 21. Crit Rev Biochem Mol Biol. 2017. PMID: 28427283 Free PMC article. Review.
-
Break-Induced Replication: The Where, The Why, and The How.Trends Genet. 2018 Jul;34(7):518-531. doi: 10.1016/j.tig.2018.04.002. Epub 2018 May 4. Trends Genet. 2018. PMID: 29735283 Free PMC article. Review.
-
Migrating bubble during break-induced replication drives conservative DNA synthesis.Nature. 2013 Oct 17;502(7471):389-92. doi: 10.1038/nature12584. Epub 2013 Sep 11. Nature. 2013. PMID: 24025772 Free PMC article.
-
Break-induced replication mechanisms in yeast and mammals.Curr Opin Genet Dev. 2021 Dec;71:163-170. doi: 10.1016/j.gde.2021.08.002. Epub 2021 Sep 1. Curr Opin Genet Dev. 2021. PMID: 34481360 Free PMC article. Review.
-
Investigation of Break-Induced Replication in Yeast.Methods Mol Biol. 2021;2153:307-328. doi: 10.1007/978-1-0716-0644-5_22. Methods Mol Biol. 2021. PMID: 32840789 Free PMC article.
Cited by
-
Consequences of telomere replication failure: the other end-replication problem.Trends Biochem Sci. 2022 Jun;47(6):506-517. doi: 10.1016/j.tibs.2022.03.013. Epub 2022 Apr 16. Trends Biochem Sci. 2022. PMID: 35440402 Free PMC article. Review.
-
Srs2 and Pif1 as Model Systems for Understanding Sf1a and Sf1b Helicase Structure and Function.Genes (Basel). 2021 Aug 26;12(9):1319. doi: 10.3390/genes12091319. Genes (Basel). 2021. PMID: 34573298 Free PMC article. Review.
-
Rdh54 stabilizes Rad51 at displacement loop intermediates to regulate genetic exchange between chromosomes.PLoS Genet. 2022 Sep 13;18(9):e1010412. doi: 10.1371/journal.pgen.1010412. eCollection 2022 Sep. PLoS Genet. 2022. PMID: 36099310 Free PMC article.
-
Helicase HELQ: Molecular Characters Fit for DSB Repair Function.Int J Mol Sci. 2024 Aug 8;25(16):8634. doi: 10.3390/ijms25168634. Int J Mol Sci. 2024. PMID: 39201320 Free PMC article. Review.
-
Protein degradation: expanding the toolbox to restrain cancer drug resistance.J Hematol Oncol. 2023 Jan 24;16(1):6. doi: 10.1186/s13045-023-01398-5. J Hematol Oncol. 2023. PMID: 36694209 Free PMC article. Review.
References
-
- Hoeijmakers JH. 2009. DNA damage, aging, and cancer. N Engl J Med 361: 1475–85 - PubMed
-
- Friedberg EC, Friedberg EC. 2006. DNA repair and mutagenesis. Washington, D.C.: ASM Press. xxix, 1118 p. pp.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

