Evasion of Intracellular DNA Sensing by Human Herpesviruses
- PMID: 33791247
- PMCID: PMC8005619
- DOI: 10.3389/fcimb.2021.647992
Evasion of Intracellular DNA Sensing by Human Herpesviruses
Abstract
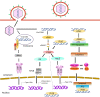
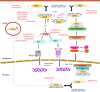
Sensing of viral constituents is the first and critical step in the host innate immune defense against viruses. In mammalian cells, there are a variety of pathogen recognition receptors (PRRs) that detect diverse pathogen-associated molecular patterns (PAMPs) including viral RNA and DNA. In the past decade, a number of host DNA sensors have been discovered and the underlying sensing mechanisms have been elucidated. Herpesviruses belong to a large family of enveloped DNA viruses. They are successful pathogens whose elaborate immune evasion mechanisms contribute to high prevalence of infection among their hosts. The three subfamilies of herpesviruses have all been found to employ diverse and overlapping strategies to interfere with host DNA sensing. These strategies include masking viral DNA or the DNA sensor, degradation of the DNA sensor, and post-transcriptional modification of the DNA sensor or its adaptor protein. In this review, we will discuss the current state of our knowledge on how human herpesviruses use these strategies to evade DNA-induced immune responses. Comprehensive understanding of herpesvirus immune-evasion mechanisms will aid in the development of vaccines and antivirals for herpesvirus-associated diseases.
Keywords: DNA sensing; cGAS; herpesvirus; innate immune response; viral evasion.
Copyright © 2021 Bhowmik and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Coevolution pays off: Herpesviruses have the license to escape the DNA sensing pathway.Med Microbiol Immunol. 2019 Aug;208(3-4):495-512. doi: 10.1007/s00430-019-00582-0. Epub 2019 Feb 25. Med Microbiol Immunol. 2019. PMID: 30805724 Review.
-
Recognition of herpesviruses by the innate immune system.Nat Rev Immunol. 2011 Feb;11(2):143-54. doi: 10.1038/nri2937. Nat Rev Immunol. 2011. PMID: 21267015 Free PMC article. Review.
-
Viral evasion of intracellular DNA and RNA sensing.Nat Rev Microbiol. 2016 Jun;14(6):360-73. doi: 10.1038/nrmicro.2016.45. Epub 2016 May 13. Nat Rev Microbiol. 2016. PMID: 27174148 Free PMC article. Review.
-
KSHV strategies for host dsDNA sensing machinery.Virol Sin. 2016 Dec;31(6):466-471. doi: 10.1007/s12250-016-3877-3. Epub 2016 Dec 5. Virol Sin. 2016. PMID: 27933565 Free PMC article. Review.
-
Innate immune response and viral interference strategies developed by human herpesviruses.Biochem Pharmacol. 2010 Dec 15;80(12):1955-72. doi: 10.1016/j.bcp.2010.07.001. Epub 2010 Jul 8. Biochem Pharmacol. 2010. PMID: 20620129 Review.
Cited by
-
Structural basis of higher order oligomerization of KSHV inhibitor of cGAS.Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2200285119. doi: 10.1073/pnas.2200285119. Epub 2022 Aug 8. Proc Natl Acad Sci U S A. 2022. PMID: 35939686 Free PMC article.
-
Teratorn and its relatives - a cross-point of distinct mobile elements, transposons and viruses.Front Vet Sci. 2023 Apr 28;10:1158023. doi: 10.3389/fvets.2023.1158023. eCollection 2023. Front Vet Sci. 2023. PMID: 37187934 Free PMC article. Review.
-
Function and regulation of cGAS-STING signaling in infectious diseases.Front Immunol. 2023 Feb 6;14:1130423. doi: 10.3389/fimmu.2023.1130423. eCollection 2023. Front Immunol. 2023. PMID: 36825026 Free PMC article. Review.
-
In Vitro Cleavage Assay to Characterize DENV NS2B3 Antagonism of cGAS.Methods Mol Biol. 2025;2854:153-170. doi: 10.1007/978-1-0716-4108-8_16. Methods Mol Biol. 2025. PMID: 39192127
-
Flavonoids Target Human Herpesviruses That Infect the Nervous System: Mechanisms of Action and Therapeutic Insights.Viruses. 2022 Mar 13;14(3):592. doi: 10.3390/v14030592. Viruses. 2022. PMID: 35336999 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

