Methylobacterium ajmalii sp. nov., Isolated From the International Space Station
- PMID: 33790880
- PMCID: PMC8005752
- DOI: 10.3389/fmicb.2021.639396
Methylobacterium ajmalii sp. nov., Isolated From the International Space Station
Abstract
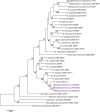
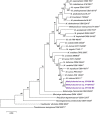
Four strains belonging to the family of Methylobacteriaceae were isolated from different locations on the International Space Station (ISS) across two consecutive flights. Of these, three were identified as Gram-negative, rod-shaped, catalase-positive, oxidase-positive, motile bacteria, designated as IF7SW-B2T, IIF1SW-B5, and IIF4SW-B5, whereas the fourth was identified as Methylorubrum rhodesianum. The sequence similarity of these three ISS strains, designated as IF7SW-B2T, IIF1SW-B5, and IIF4SW-B5, was <99.4% for 16S rRNA genes and <97.3% for gyrB gene, with the closest being Methylobacterium indicum SE2.11T. Furthermore, the multi-locus sequence analysis placed these three ISS strains in the same clade of M. indicum. The average nucleotide identity (ANI) values of these three ISS strains were <93% and digital DNA-DNA hybridization (dDDH) values were <46.4% with any described Methylobacterium species. Based on the ANI and dDDH analyses, these three ISS strains were considered as novel species belonging to the genus Methylobacterium. The three ISS strains showed 100% ANI similarity and dDDH values with each other, indicating that these three ISS strains, isolated during various flights and from different locations, belong to the same species. These three ISS strains were found to grow optimally at temperatures from 25 to 30°C, pH 6.0 to 8.0, and NaCl 0 to 1%. Phenotypically, these three ISS strains resemble M. aquaticum and M. terrae since they assimilate similar sugars as sole carbon substrate when compared to other Methylobacterium species. Fatty acid analysis showed that the major fatty acid produced by the ISS strains are C18 : 1-ω7c and C18 : 1-ω6c. The predominant quinone was ubiquinone 10, and the major polar lipids were diphosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, and an unidentified lipid. Therefore, based on genomic, phylogenetic, biochemical, and fatty acid analyses, strains IF7SW-B2T, IIF1SW-B5, and IIF4SW-B5, are assigned to a novel species within the genus Methylobacterium, and the name Methylobacterium ajmalii sp. nov. is proposed. The type strain is IF7SW-B2T (NRRL B-65601T and LMG 32165T).
Keywords: ANI; Methylobacterium; international space station (ISS); polyphasic taxomony; whole genome sequencing.
Copyright © 2021 Bijlani, Singh, Eedara, Podile, Mason, Wang and Venkateswaran.
Conflict of interest statement
The author(s) declare that there are no conflicts of interest. This manuscript was prepared as an account of work sponsored by NASA, an agency of the US Government. The US Government, NASA, California Institute of Technology, Jet Propulsion Laboratory, and their employees make no warranty, expressed or implied, or assume any liability or responsibility for the accuracy, completeness, or usefulness of information, apparatus, product, or process disclosed in this manuscript, or represents that its use would not infringe upon privately held rights. The use of, and references to any commercial product, process, or service does not necessarily constitute or imply endorsement, recommendation, or favoring by the U.S. Government, NASA, California Institute of Technology, or Jet Propulsion Laboratory. Views and opinions presented herein by the authors of this manuscript do not necessarily reflect those of the U.S. Government, NASA, California Institute of Technology, or Jet Propulsion Laboratory, and shall not be used for advertisements or product endorsements.
Figures


Similar articles
-
Methylobacterium indicum sp. nov., a facultative methylotrophic bacterium isolated from rice seed.Syst Appl Microbiol. 2016 Feb;39(1):25-32. doi: 10.1016/j.syapm.2015.12.006. Epub 2015 Dec 29. Syst Appl Microbiol. 2016. PMID: 26776106
-
Paraburkholderia acidiphila sp. nov., Paraburkholderia acidisoli sp. nov. and Burkholderia guangdongensis sp. nov., isolated from forest soil, and reclassification of Burkholderia ultramafica as Paraburkholderia ultramafica comb. nov.Int J Syst Evol Microbiol. 2019 Jun;71(3). doi: 10.1099/ijsem.0.004690. Epub 2021 Feb 8. Int J Syst Evol Microbiol. 2019. PMID: 33555242
-
Allorhizobium terrae sp. nov., isolated from paddy soil, and reclassification of Rhizobium oryziradicis (Zhao et al. 2017) as Allorhizobium oryziradicis comb. nov.Int J Syst Evol Microbiol. 2020 Jan;70(1):397-405. doi: 10.1099/ijsem.0.003770. Int J Syst Evol Microbiol. 2020. PMID: 31626583
-
Methylobacterium crusticola sp. nov., isolated from biological soil crusts.Int J Syst Evol Microbiol. 2020 Mar;70(3):2089-2095. doi: 10.1099/ijsem.0.004020. Int J Syst Evol Microbiol. 2020. PMID: 31995465
-
Methylobacterium oryzihabitans sp. nov., isolated from water sampled from a rice paddy field.Int J Syst Evol Microbiol. 2019 Dec;69(12):3843-3850. doi: 10.1099/ijsem.0.003693. Int J Syst Evol Microbiol. 2019. PMID: 31483751
Cited by
-
Understanding the Complexities and Changes of the Astronaut Microbiome for Successful Long-Duration Space Missions.Life (Basel). 2022 Mar 28;12(4):495. doi: 10.3390/life12040495. Life (Basel). 2022. PMID: 35454986 Free PMC article. Review.
-
A survey of Methylobacterium species and strains reveals widespread production and varying profiles of cytokinin phytohormones.BMC Microbiol. 2022 Feb 8;22(1):49. doi: 10.1186/s12866-022-02454-9. BMC Microbiol. 2022. PMID: 35135483 Free PMC article.
-
Mitigation and use of biofilms in space for the benefit of human space exploration.Biofilm. 2023 Jan 6;5:100102. doi: 10.1016/j.bioflm.2022.100102. eCollection 2023 Dec. Biofilm. 2023. PMID: 36660363 Free PMC article.
-
From genome to evolution: investigating type II methylotrophs using a pangenomic analysis.mSystems. 2024 Jun 18;9(6):e0024824. doi: 10.1128/msystems.00248-24. Epub 2024 May 2. mSystems. 2024. PMID: 38695578 Free PMC article.
-
Characterization of endophytic bacteriome diversity and associated beneficial bacteria inhabiting a macrophyte Eichhornia crassipes.Front Plant Sci. 2023 Jun 19;14:1176648. doi: 10.3389/fpls.2023.1176648. eCollection 2023. Front Plant Sci. 2023. PMID: 37404529 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

