Type I interferons affect the metabolic fitness of CD8+ T cells from patients with systemic lupus erythematosus
- PMID: 33790300
- PMCID: PMC8012390
- DOI: 10.1038/s41467-021-22312-y
Type I interferons affect the metabolic fitness of CD8+ T cells from patients with systemic lupus erythematosus
Abstract
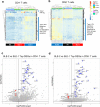
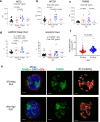
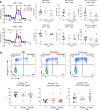
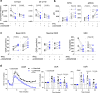
The majority of patients with systemic lupus erythematosus (SLE) have high expression of type I IFN-stimulated genes. Mitochondrial abnormalities have also been reported, but the contribution of type I IFN exposure to these changes is unknown. Here, we show downregulation of mitochondria-derived genes and mitochondria-associated metabolic pathways in IFN-High patients from transcriptomic analysis of CD4+ and CD8+ T cells. CD8+ T cells from these patients have enlarged mitochondria and lower spare respiratory capacity associated with increased cell death upon rechallenge with TCR stimulation. These mitochondrial abnormalities can be phenocopied by exposing CD8+ T cells from healthy volunteers to type I IFN and TCR stimulation. Mechanistically these 'SLE-like' conditions increase CD8+ T cell NAD+ consumption resulting in impaired mitochondrial respiration and reduced cell viability, both of which can be rectified by NAD+ supplementation. Our data suggest that type I IFN exposure contributes to SLE pathogenesis by promoting CD8+ T cell death via metabolic rewiring.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
IFNs disrupt T cell metabolism in SLE.Nat Rev Rheumatol. 2021 Jun;17(6):310. doi: 10.1038/s41584-021-00622-1. Nat Rev Rheumatol. 2021. PMID: 33907324 No abstract available.
Similar articles
-
SIT1 identifies circulating hypoactive T cells with elevated cytotoxic molecule secretion in systemic lupus erythematosus patients.Immunol Res. 2024 Aug;72(4):754-765. doi: 10.1007/s12026-024-09481-w. Epub 2024 May 1. Immunol Res. 2024. PMID: 38691318
-
Double-Edged Sword: Interleukin-2 Promotes T Regulatory Cell Differentiation but Also Expands Interleukin-13- and Interferon-γ-Producing CD8+ T Cells via STAT6-GATA-3 Axis in Systemic Lupus Erythematosus.Front Immunol. 2021 Mar 2;12:635531. doi: 10.3389/fimmu.2021.635531. eCollection 2021. Front Immunol. 2021. PMID: 33763079 Free PMC article.
-
Hypomethylation of STAT1 and HLA-DRB1 is associated with type-I interferon-dependent HLA-DRB1 expression in lupus CD8+ T cells.Ann Rheum Dis. 2019 Apr;78(4):519-528. doi: 10.1136/annrheumdis-2018-214323. Epub 2019 Jan 23. Ann Rheum Dis. 2019. PMID: 30674474 Free PMC article.
-
A possible pathogenic role of CD8+ T cells and their derived cytokine, IL-16, in SLE.Autoimmunity. 2000;33(1):37-44. doi: 10.3109/08916930108994108. Autoimmunity. 2000. PMID: 11204252 Review.
-
Mitochondrial bioenergetic changes in systemic lupus erythematosus immune cell subsets: Contributions to pathogenesis and clinical applications.Lupus. 2023 Apr;32(5):603-611. doi: 10.1177/09612033231164635. Epub 2023 Mar 13. Lupus. 2023. PMID: 36914582 Free PMC article. Review.
Cited by
-
The role of mitochondria in rheumatic diseases.Nat Rev Rheumatol. 2022 Nov;18(11):621-640. doi: 10.1038/s41584-022-00834-z. Epub 2022 Sep 29. Nat Rev Rheumatol. 2022. PMID: 36175664 Review.
-
Cell type-specific mechanistic target of rapamycin-dependent distortion of autophagy pathways in lupus nephritis.Transl Res. 2022 Jul;245:55-81. doi: 10.1016/j.trsl.2022.03.004. Epub 2022 Mar 12. Transl Res. 2022. PMID: 35288362 Free PMC article. Review.
-
Multi-omics identify falling LRRC15 as a COVID-19 severity marker and persistent pro-thrombotic signals in convalescence.Nat Commun. 2022 Dec 15;13(1):7775. doi: 10.1038/s41467-022-35454-4. Nat Commun. 2022. PMID: 36522333 Free PMC article.
-
COX5A as a potential biomarker for disease activity and organ damage in lupus.Clin Exp Med. 2023 Dec;23(8):4745-4756. doi: 10.1007/s10238-023-01215-w. Epub 2023 Oct 27. Clin Exp Med. 2023. PMID: 37891386
-
Optimal CD8+ T cell effector function requires costimulation-induced RNA-binding proteins that reprogram the transcript isoform landscape.Nat Commun. 2022 Jun 20;13(1):3540. doi: 10.1038/s41467-022-31228-0. Nat Commun. 2022. PMID: 35725727 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

