Novel circular RNA circSOBP governs amoeboid migration through the regulation of the miR-141-3p/MYPT1/p-MLC2 axis in prostate cancer
- PMID: 33784000
- PMCID: PMC8002909
- DOI: 10.1002/ctm2.360
Novel circular RNA circSOBP governs amoeboid migration through the regulation of the miR-141-3p/MYPT1/p-MLC2 axis in prostate cancer
Abstract
Background: Metastatic prostate cancer is a fatal disease despite multiple new approvals in recent years. Recent studies revealed that circular RNAs (circRNAs) can be involved in cancer metastasis. Defining the role of circRNAs in prostate cancer metastasis and discovering therapeutic targets that block cancer metastasis is of great significance for the treatment of prostate cancer.
Methods: The circSOBP levels in prostate cancer (PCa) were determined by qRT-PCR. We evaluated the function of circSOBP using a transwell assay and nude mice lung metastasis models. Immunofluorescence assay and electron microscopic assay were applied to determine the phenotypes of prostate cancer cells' migration. We used fluorescence in situ hybridization assay to determine the localization of RNAs. Dual luciferase and rescue assays were applied to verify the interactions between circSOBP, miR-141-3p, MYPT1, and phosphomyosin light chain (p-MLC2).
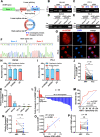
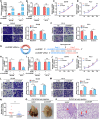
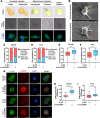
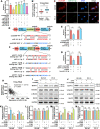
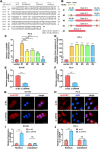
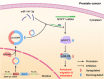
Results: We observed that circSOBP level was significantly lower in PCa specimens compared with adjacent noncancerous prostate specimens, and was correlated with the grade group of PCa. Overexpression of circSOBP suppressed PCa migration and invasion in vitro and metastasis in vivo. CircSOBP depletion increased migration and invasion and induced amoeboid migration of PCa cells. Mechanistically, circSOBP bound miR-141-3p and regulated the MYPT1/p-MLC2 axis. Moreover, the depletion of MYPT1 reversed the inhibitory effect of circSOBP on the migration and invasion of PCa cells. Complementary intronic Alu elements induced but were not necessary for the formation of circSOBP. The nuclear export of circSOBP was mediated by URH49.
Conclusion: Our results suggest that circSOBP suppresses amoeboid migration of PCa cells and inhibits migration and invasion through sponging miR-141-3p and regulating the MYPT1/p-MLC2 axis.
Keywords: amoeboid migration; circRNA; circSOBP; metastasis; prostate cancer.
© 2021 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
circSOBP Inhibits Bladder Cancer Proliferation and Metastasis by Regulating the miR-200a-3p/PTEN Axis and Participating in the Immune Response.Cell Transplant. 2023 Jan-Dec;32:9636897231165874. doi: 10.1177/09636897231165874. Cell Transplant. 2023. PMID: 37026617 Free PMC article.
-
MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer.Mol Cancer. 2017 Feb 27;16(1):48. doi: 10.1186/s12943-017-0615-x. Mol Cancer. 2017. Retraction in: Mol Cancer. 2023 Mar 20;22(1):56. doi: 10.1186/s12943-023-01763-5 PMID: 28241827 Free PMC article. Retracted.
-
Circular RNA EPHA3 suppresses progression and metastasis in prostate cancer through the miR-513a-3p/BMP2 axis.J Transl Med. 2023 Apr 28;21(1):288. doi: 10.1186/s12967-023-04132-4. J Transl Med. 2023. PMID: 37118847 Free PMC article.
-
Role of Circular RNAs in Prostate Cancer.Curr Med Chem. 2024;31(29):4640-4656. doi: 10.2174/0929867330666230531095850. Curr Med Chem. 2024. PMID: 37259936 Review.
-
Plasticity of cancer invasion and energy metabolism.Trends Cell Biol. 2023 May;33(5):388-402. doi: 10.1016/j.tcb.2022.09.009. Epub 2022 Oct 31. Trends Cell Biol. 2023. PMID: 36328835 Free PMC article. Review.
Cited by
-
The short inverted repeats-induced circEXOC6B inhibits prostate cancer metastasis by enhancing the binding of RBMS1 and HuR.Mol Ther. 2023 Jun 7;31(6):1705-1721. doi: 10.1016/j.ymthe.2022.08.006. Epub 2022 Aug 15. Mol Ther. 2023. PMID: 35974702 Free PMC article.
-
Interactions between circRNAs and miR-141 in Cancer: From Pathogenesis to Diagnosis and Therapy.Int J Mol Sci. 2023 Jul 24;24(14):11861. doi: 10.3390/ijms241411861. Int J Mol Sci. 2023. PMID: 37511619 Free PMC article. Review.
-
MicroRNAs Present in Malignant Pleural Fluid Increase the Migration of Normal Mesothelial Cells In Vitro and May Help Discriminate between Benign and Malignant Effusions.Int J Mol Sci. 2023 Sep 13;24(18):14022. doi: 10.3390/ijms241814022. Int J Mol Sci. 2023. PMID: 37762343 Free PMC article.
-
ZEB1-mediated biogenesis of circNIPBL sustains the metastasis of bladder cancer via Wnt/β-catenin pathway.J Exp Clin Cancer Res. 2023 Aug 2;42(1):191. doi: 10.1186/s13046-023-02757-3. J Exp Clin Cancer Res. 2023. PMID: 37528489 Free PMC article.
-
The role of circular RNA during the urological cancer metastasis: exploring regulatory mechanisms and potential therapeutic targets.Cancer Metastasis Rev. 2024 Sep;43(3):1055-1074. doi: 10.1007/s10555-024-10182-x. Epub 2024 Apr 1. Cancer Metastasis Rev. 2024. PMID: 38558156 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
