Circular RNAs: Their Role in the Pathogenesis and Orchestration of Breast Cancer
- PMID: 33777954
- PMCID: PMC7991790
- DOI: 10.3389/fcell.2021.647736
Circular RNAs: Their Role in the Pathogenesis and Orchestration of Breast Cancer
Abstract
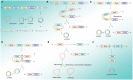
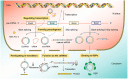
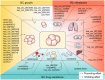
As one of the most frequently occurring malignancies in women, breast cancer (BC) is still an enormous threat to women all over the world. The high mortality rates in BC patients are associated with BC recurrence, metastatic progression to distant organs, and therapeutic resistance. Circular RNAs (circRNAs), belonging to the non-coding RNAs (ncRNAs), are connected end to end to form covalently closed single-chain circular molecules. CircRNAs are widely found in different species and a variety of human cells, with the features of diversity, evolutionary conservation, stability, and specificity. CircRNAs are emerging important participators in multiple diseases, including cardiovascular disease, inflammation, and cancer. Recent studies have shown that circRNAs are involved in BC progress by regulating gene expression at the transcriptional or post-transcriptional level via binding to miRNAs then inhibiting their function, suggesting that circRNAs may be potential targets for early diagnosis, treatment, and prognosis of BC. Herein, in this article, we have reviewed and summarized the current studies about the biogenesis, features, and functions of circRNAs. More importantly, we emphatically elucidate the pivotal functions and mechanisms of circRNAs in BC growth, metastasis, diagnosis, and drug resistance. Deciphering the complex networks, especially the circRNA-miRNA target gene axis, will endow huge potentials in developing therapeutic strategies for combating BC.
Keywords: breast cancer; circular RNA; diagnosis; drug resistance; metastasis; microRNA sponge.
Copyright © 2021 He, Xu, Hu, Tan, Wang, Wang, Zhao, Yi, Xiong, Lv, Wu, Li, Wu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Circular RNA and its potential diagnostic and therapeutic values in breast cancer.Mol Biol Rep. 2024 Feb 1;51(1):258. doi: 10.1007/s11033-023-09172-z. Mol Biol Rep. 2024. PMID: 38302635 Review.
-
In silico analysis and comprehensive review of circular-RNA regulatory roles in breast diseases; a step-toward non-coding RNA precision.Pathol Res Pract. 2024 Nov;263:155651. doi: 10.1016/j.prp.2024.155651. Epub 2024 Oct 19. Pathol Res Pract. 2024. PMID: 39454476 Review.
-
Tumor-Derived Exosomal Non-Coding RNAs: The Emerging Mechanisms and Potential Clinical Applications in Breast Cancer.Front Oncol. 2021 Oct 11;11:738945. doi: 10.3389/fonc.2021.738945. eCollection 2021. Front Oncol. 2021. PMID: 34707990 Free PMC article. Review.
-
CircRNAs: Emerging Bladder Cancer Biomarkers and Targets.Front Oncol. 2021 Jan 8;10:606485. doi: 10.3389/fonc.2020.606485. eCollection 2020. Front Oncol. 2021. PMID: 33489913 Free PMC article. Review.
-
Circular RNAs in cancer: novel insights into origins, properties, functions and implications.Am J Cancer Res. 2015 Jan 15;5(2):472-80. eCollection 2015. Am J Cancer Res. 2015. PMID: 25973291 Free PMC article. Review.
Cited by
-
Circular RNA expression alterations and function prediction in OSA-induced pancreatic injury in mice: New insights into pathogenesis.PLoS One. 2023 Apr 14;18(4):e0284459. doi: 10.1371/journal.pone.0284459. eCollection 2023. PLoS One. 2023. PMID: 37058537 Free PMC article.
-
Stem Cell-Derived Extracellular Vesicles: Promising Therapeutic Opportunities for Diabetic Wound Healing.Int J Nanomedicine. 2024 May 17;19:4357-4375. doi: 10.2147/IJN.S461342. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38774027 Free PMC article. Review.
-
The Emerging Functions of Circular RNAs in Bladder Cancer.Cancers (Basel). 2021 Sep 15;13(18):4618. doi: 10.3390/cancers13184618. Cancers (Basel). 2021. PMID: 34572845 Free PMC article. Review.
-
Downregulated circPOKE promotes breast cancer metastasis through activation of the USP10-Snail axis.Oncogene. 2023 Oct;42(44):3236-3251. doi: 10.1038/s41388-023-02823-2. Epub 2023 Sep 16. Oncogene. 2023. PMID: 37717099
-
circRNAs in drug resistance of breast cancer.Oncol Res. 2023 Jan 31;30(4):157-172. doi: 10.32604/or.2022.027547. eCollection 2022. Oncol Res. 2023. PMID: 37304411 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

