Holomycin, an Antibiotic Secondary Metabolite, Is Required for Biofilm Formation by the Native Producer Photobacterium galatheae S2753
- PMID: 33771780
- PMCID: PMC8208159
- DOI: 10.1128/AEM.00169-21
Holomycin, an Antibiotic Secondary Metabolite, Is Required for Biofilm Formation by the Native Producer Photobacterium galatheae S2753
Abstract
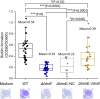
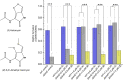
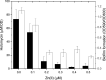
While the effects of antibiotics on microorganisms are widely studied, it remains less well understood how antibiotics affect the physiology of the native producing organisms. Here, using a marine bacterium, Photobacterium galatheae S2753, that produces the antibiotic holomycin, we generated a holomycin-deficient strain by in-frame deletion of hlmE, the core gene responsible for holomycin production. Mass spectrometry analysis of cell extracts confirmed that the ΔhlmE strain did not produce holomycin and that the mutant was devoid of antibacterial activity. Biofilm formation of the ΔhlmE strain was significantly reduced compared to that of wild-type S2753 and was restored in an hlmE complementary mutant. Consistent with this, exogenous holomycin, but not its dimethylated and less antibacterial derivative, S,S'-dimethyl holomycin, restored the biofilm formation of the ΔhlmE strain. Furthermore, zinc starvation was found to be essential for both holomycin production and biofilm formation of S2753, although the molecular mechanism remains elusive. Collectively, these data suggest that holomycin promotes biofilm formation of S2753 via its ene-disulfide group. Lastly, the addition of holomycin at subinhibitory concentrations also enhanced the biofilms of four other Vibrionaceae strains. P. galatheae likely gains an ecological advantage from producing holomycin as both an antibiotic and a biofilm stimulator, which facilitates nutrition acquisition and protects P. galatheae from environmental stresses. Studying the function of antibiotic compounds in the native producer will shed light on their roles in nature and could point to novel bioprospecting strategies.IMPORTANCE Despite the societal impact of antibiotics, their ecological functions remain elusive and have mostly been studied by exposing nonproducing bacteria to subinhibitory concentrations. Here, we studied the effects of the antibiotic holomycin on its native producer, Photobacterium galatheae S2753, a Vibrionaceae bacterium. Holomycin provides a distinct advantage to S2753 both as an antibiotic and by enhancing biofilm formation in the producer. Vibrionaceae species successfully thrive in global marine ecosystems, where they play critical ecological roles as free-living, symbiotic, or pathogenic bacteria. Genome mining has demonstrated that many have the potential to produce several bioactive compounds, including P. galatheae To unravel the contribution of the microbial metabolites to the development of marine microbial ecosystems, better insight into the function of these compounds in the producing organisms is needed. Our finding provides a model to pursue this and highlights the ecological importance of antibiotics to the fitness of the producing organisms.
Keywords: Photobacterium galatheae; biofilm; biosynthetic gene cluster; holomycin; secondary metabolites.
Copyright © 2021 American Society for Microbiology.
Figures

Similar articles
-
Solonamides, a Group of Cyclodepsipeptides, Influence Motility in the Native Producer Photobacterium galatheae S2753.Appl Environ Microbiol. 2022 Sep 13;88(17):e0110522. doi: 10.1128/aem.01105-22. Epub 2022 Aug 24. Appl Environ Microbiol. 2022. PMID: 36000852 Free PMC article.
-
Enhancement of antibiotic production by co-cultivation of two antibiotic producing marine Vibrionaceae strains.FEMS Microbiol Ecol. 2021 Mar 31;97(4):fiab041. doi: 10.1093/femsec/fiab041. FEMS Microbiol Ecol. 2021. PMID: 33693627
-
Growth on Chitin Impacts the Transcriptome and Metabolite Profiles of Antibiotic-Producing Vibrio coralliilyticus S2052 and Photobacterium galatheae S2753.mSystems. 2017 Jan 3;2(1):e00141-16. doi: 10.1128/mSystems.00141-16. eCollection 2017 Jan-Feb. mSystems. 2017. PMID: 28066819 Free PMC article.
-
Holomycin, a dithiolopyrrolone compound produced by Streptomyces clavuligerus.Appl Microbiol Biotechnol. 2014 Feb;98(3):1023-30. doi: 10.1007/s00253-013-5410-z. Epub 2013 Dec 10. Appl Microbiol Biotechnol. 2014. PMID: 24323287 Review.
-
The biology and the importance of Photobacterium species.Appl Microbiol Biotechnol. 2017 Jun;101(11):4371-4385. doi: 10.1007/s00253-017-8300-y. Epub 2017 May 11. Appl Microbiol Biotechnol. 2017. PMID: 28497204 Review.
Cited by
-
Solonamides, a Group of Cyclodepsipeptides, Influence Motility in the Native Producer Photobacterium galatheae S2753.Appl Environ Microbiol. 2022 Sep 13;88(17):e0110522. doi: 10.1128/aem.01105-22. Epub 2022 Aug 24. Appl Environ Microbiol. 2022. PMID: 36000852 Free PMC article.
-
The potential to produce tropodithietic acid by Phaeobacter inhibens affects the assembly of microbial biofilm communities in natural seawater.NPJ Biofilms Microbiomes. 2023 Mar 23;9(1):12. doi: 10.1038/s41522-023-00379-3. NPJ Biofilms Microbiomes. 2023. PMID: 36959215 Free PMC article.
-
Role is in the eye of the beholder-the multiple functions of the antibacterial compound tropodithietic acid produced by marine Rhodobacteraceae.FEMS Microbiol Rev. 2022 May 6;46(3):fuac007. doi: 10.1093/femsre/fuac007. FEMS Microbiol Rev. 2022. PMID: 35099011 Free PMC article.
-
Tropodithietic Acid, a Multifunctional Antimicrobial, Facilitates Adaption and Colonization of the Producer, Phaeobacter piscinae.mSphere. 2023 Feb 21;8(1):e0051722. doi: 10.1128/msphere.00517-22. Epub 2023 Jan 9. mSphere. 2023. PMID: 36622251 Free PMC article.
-
Distribution and comparative genomic analysis of antimicrobial gene clusters found in Pantoea.Front Microbiol. 2024 Aug 14;15:1416674. doi: 10.3389/fmicb.2024.1416674. eCollection 2024. Front Microbiol. 2024. PMID: 39206372 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

