Complete chloroplast genome sequence and phylogenetic analysis of winter oil rapeseed (Brassica rapa L.)
- PMID: 33763561
- PMCID: PMC7954489
- DOI: 10.1080/23802359.2020.1860697
Complete chloroplast genome sequence and phylogenetic analysis of winter oil rapeseed (Brassica rapa L.)
Abstract
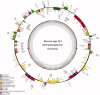
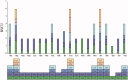
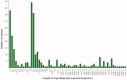
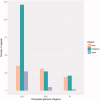
Winter oil rapeseed '18 R-1' (Brassica rapa L.) is a new variety that can survive in northern China where the extreme low temperature is -20 °C to -32 °C. It is different from traditional B. rapa and Brassica napus. In this study, the complete chloroplast (cp) genome of '18 R-1' was sequenced and analyzed to assess the genetic relationship. The size of cp genome is 153,494 bp, including one large single copy (LSC) region of 83,280 bp and one small single copy (SSC) region of 17,776 bp, separated by two inverted repeat (IR) regions of 26,219 bp. The GC content of the whole genome is 36.35%, while those of LSC, SSC, and IR are 34.12%, 29.20%, and 42.32%, respectively. The cp genome encodes 132 genes, including 87 protein-coding genes, eight rRNA genes, and 37 tRNA genes. In repeat structure analysis, 288 simple sequence repeats (SSRs) were identified. Cp genome of '18 R-1' was closely related to Brassica chinensis, B. rapa and Brassica pekinesis.
Keywords: SSRs; Winter oil rapeseed; chloroplast genome; phylogeny.
© 2021 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
The complete chloroplast genome of Brassica rapa var. purpuraria (L.H.Bailey) Kitam 1950 and its phylogenetic analysis.Mitochondrial DNA B Resour. 2024 Jan 24;9(1):143-147. doi: 10.1080/23802359.2024.2305403. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38274856 Free PMC article.
-
The Complete Chloroplast Genome of a Key Ancestor of Modern Roses, Rosa chinensis var. spontanea, and a Comparison with Congeneric Species.Molecules. 2018 Feb 12;23(2):389. doi: 10.3390/molecules23020389. Molecules. 2018. PMID: 29439505 Free PMC article.
-
Complete chloroplast genome sequence of turnip (Brassica rapa. ssp. rapa): genome structure and phylogenetic analysis.Mitochondrial DNA B Resour. 2020 Oct 9;5(3):3555-3557. doi: 10.1080/23802359.2020.1829124. Mitochondrial DNA B Resour. 2020. PMID: 33458239 Free PMC article.
-
The complete chloroplast genome sequence of Indian mustard (Brassica juncea L.).Mitochondrial DNA A DNA Mapp Seq Anal. 2016 Nov;27(6):4622-4623. doi: 10.3109/19401736.2015.1101586. Epub 2015 Dec 28. Mitochondrial DNA A DNA Mapp Seq Anal. 2016. PMID: 26708222
-
The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms.Gene. 2013 Oct 10;528(2):120-31. doi: 10.1016/j.gene.2013.07.037. Epub 2013 Jul 27. Gene. 2013. PMID: 23900198
Cited by
-
The Acinetobacter baumannii model can explain the role of small non-coding RNAs as potential mediators of host-pathogen interactions.Front Mol Biosci. 2022 Dec 21;9:1088783. doi: 10.3389/fmolb.2022.1088783. eCollection 2022. Front Mol Biosci. 2022. PMID: 36619166 Free PMC article. Review.
-
The complete plastome of Acalypha australis (Euphorbiaceae) and its phylogenetic analysis.Mitochondrial DNA B Resour. 2024 May 17;9(5):636-640. doi: 10.1080/23802359.2023.2294891. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38770147 Free PMC article.
-
The complete chloroplast genome of Brassica rapa var. purpuraria (L.H.Bailey) Kitam 1950 and its phylogenetic analysis.Mitochondrial DNA B Resour. 2024 Jan 24;9(1):143-147. doi: 10.1080/23802359.2024.2305403. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 38274856 Free PMC article.
References
-
- Dongmei Z, Renzhi Z, Wancang S, Jun Z, Heling W.. 2014. Evaluation of the suitability and influencing factors of winter rapeseed planting in Gansu Province. Chin J Ecoagric. 22:697–704.
-
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
