Virtual 2-D map of the fungal proteome
- PMID: 33758316
- PMCID: PMC7988114
- DOI: 10.1038/s41598-021-86201-6
Virtual 2-D map of the fungal proteome
Abstract
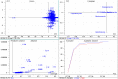
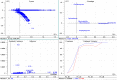
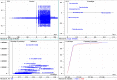
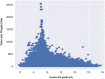
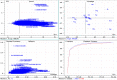
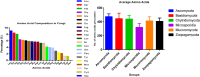
The molecular weight and isoelectric point (pI) of the proteins plays important role in the cell. Depending upon the shape, size, and charge, protein provides its functional role in different parts of the cell. Therefore, understanding to the knowledge of their molecular weight and charges is (pI) is very important. Therefore, we conducted a proteome-wide analysis of protein sequences of 689 fungal species (7.15 million protein sequences) and construct a virtual 2-D map of the fungal proteome. The analysis of the constructed map revealed the presence of a bimodal distribution of fungal proteomes. The molecular mass of individual fungal proteins ranged from 0.202 to 2546.166 kDa and the predicted isoelectric point (pI) ranged from 1.85 to 13.759 while average molecular weight of fungal proteome was 50.98 kDa. A non-ribosomal peptide synthase (RFU80400.1) found in Trichoderma arundinaceum was identified as the largest protein in the fungal kingdom. The collective fungal proteome is dominated by the presence of acidic rather than basic pI proteins and Leu is the most abundant amino acid while Cys is the least abundant amino acid. Aspergillus ustus encodes the highest percentage (76.62%) of acidic pI proteins while Nosema ceranae was found to encode the highest percentage (66.15%) of basic pI proteins. Selenocysteine and pyrrolysine amino acids were not found in any of the analysed fungal proteomes. Although the molecular weight and pI of the protein are of enormous important to understand their functional roles, the amino acid compositions of the fungal protein will enable us to understand the synonymous codon usage in the fungal kingdom. The small peptides identified during the study can provide additional biotechnological implication.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Virtual 2D mapping of the viral proteome reveals host-specific modality distribution of molecular weight and isoelectric point.Sci Rep. 2021 Oct 28;11(1):21291. doi: 10.1038/s41598-021-00797-3. Sci Rep. 2021. PMID: 34711905 Free PMC article.
-
The molecular mass and isoelectric point of plant proteomes.BMC Genomics. 2019 Aug 5;20(1):631. doi: 10.1186/s12864-019-5983-8. BMC Genomics. 2019. PMID: 31382875 Free PMC article.
-
Decoding the Virtual 2D Map of the Chloroplast Proteomes.Biol Proced Online. 2022 Dec 13;24(1):23. doi: 10.1186/s12575-022-00186-8. Biol Proced Online. 2022. PMID: 36513972 Free PMC article.
-
Exploring Trichoderma and Aspergillus secretomes: Proteomics approaches for the identification of enzymes of biotechnological interest.Enzyme Microb Technol. 2018 Feb;109:1-10. doi: 10.1016/j.enzmictec.2017.08.007. Epub 2017 Aug 24. Enzyme Microb Technol. 2018. PMID: 29224620 Review.
-
Fungal proteomics: from identification to function.FEMS Microbiol Lett. 2011 Aug;321(1):1-9. doi: 10.1111/j.1574-6968.2011.02292.x. Epub 2011 May 13. FEMS Microbiol Lett. 2011. PMID: 21517945 Review.
Cited by
-
Virtual 2D mapping of the viral proteome reveals host-specific modality distribution of molecular weight and isoelectric point.Sci Rep. 2021 Oct 28;11(1):21291. doi: 10.1038/s41598-021-00797-3. Sci Rep. 2021. PMID: 34711905 Free PMC article.
-
FungiProteomeDB: a database for the molecular weight and isoelectric points of the fungal proteomes.Database (Oxford). 2023 Mar 16;2023:baad004. doi: 10.1093/database/baad004. Database (Oxford). 2023. PMID: 36929177 Free PMC article.
-
Proteome-pI 2.0: proteome isoelectric point database update.Nucleic Acids Res. 2022 Jan 7;50(D1):D1535-D1540. doi: 10.1093/nar/gkab944. Nucleic Acids Res. 2022. PMID: 34718696 Free PMC article.
-
PlantMWpIDB: a database for the molecular weight and isoelectric points of the plant proteomes.Sci Rep. 2022 May 6;12(1):7421. doi: 10.1038/s41598-022-11077-z. Sci Rep. 2022. PMID: 35523906 Free PMC article.
-
Virtual 2D map of cyanobacterial proteomes.PLoS One. 2022 Oct 3;17(10):e0275148. doi: 10.1371/journal.pone.0275148. eCollection 2022. PLoS One. 2022. PMID: 36190972 Free PMC article.
References
-
- Vincent D, Zivy M. Plant proteome responses to abiotic stress. In: Šamaj J, Thelen JJ, editors. Plant Proteomics. Springer; 2007. pp. 346–364.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

