The subcellular redistribution of NLRC5 promotes angiogenesis via interacting with STAT3 in endothelial cells
- PMID: 33754073
- PMCID: PMC7977449
- DOI: 10.7150/thno.54473
The subcellular redistribution of NLRC5 promotes angiogenesis via interacting with STAT3 in endothelial cells
Abstract
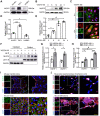
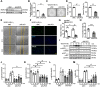
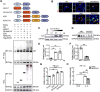
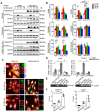
Angiogenesis is a critical step in repair of tissue injury. The pattern recognition receptors (PRRs) recognize pathogen and damage associated molecular patterns (DAMPs) during injury and achieve host defense directly. However, the role of NLR family CARD domain containing 5 (NLRC5), an important member of PPRs, beyond host defense in angiogenesis during tissue repair remains unknown. Methods:In vitro, western blot and real-time PCR (RT-PCR) were used to detect the expression of NLRC5 in endothelial cells (ECs). Immunofluorescence microscopy was used to reveal the subcellular location of NLRC5 in ECs. Cell proliferation, wound healing, tube formation assays of ECs were performed to study the role of NLRC5 in angiogenesis. By using Tie2Cre-NLRC5flox/flox mice and bone marrow transplantation studies, we defined an EC-specific role for NLRC5 in angiogenesis. Mechanistically, co-immunoprecipitation studies and RNA sequencing indicated that signal transducer and activator of transcription 3 (STAT3) was the target of NLRC5 in the nucleus. And Co-IP was used to verify the specific domain of NLRC5 binding with STAT3. ChIP assay determined the genes regulated by interaction of STAT3 and NLRC5. Results: Knockdown of NLRC5 in vitro or in vivo inhibited pathological angiogenesis, but had no effect on physiological angiogenesis. NLRC5 was also identified to bind to STAT3 in the nucleus required the integrated death-domain and nucleotide-binding domain (DD+NACHT domain) of NLRC5. And the interaction of STAT3 and NLRC5 could enhance the transcription of angiopoietin-2 (Ang2) and cyclin D1 (CCND1) to participate in angiogenesis. Conclusions: In the ischemic microenvironment, NLRC5 protein accumulates in the nucleus of ECs and enhances STAT3 transcriptional activity for angiogenesis. These findings establish NLRC5 as a novel modulator of VEGFA signaling, providing a new target for angiogenic therapy to foster tissue regeneration.
Keywords: NLRC5; STAT3; angiogenesis; endothelial cell; signal transduction.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Yes-Associated Protein Promotes Angiogenesis via Signal Transducer and Activator of Transcription 3 in Endothelial Cells.Circ Res. 2018 Feb 16;122(4):591-605. doi: 10.1161/CIRCRESAHA.117.311950. Epub 2018 Jan 3. Circ Res. 2018. PMID: 29298775
-
Mineralocorticoid receptor negatively regulates angiogenesis through repression of STAT3 activity in endothelial cells.J Pathol. 2019 Aug;248(4):438-451. doi: 10.1002/path.5269. Epub 2019 May 6. J Pathol. 2019. PMID: 30900255
-
S-propargyl-cysteine, a novel water-soluble modulator of endogenous hydrogen sulfide, promotes angiogenesis through activation of signal transducer and activator of transcription 3.Antioxid Redox Signal. 2014 May 20;20(15):2303-16. doi: 10.1089/ars.2013.5449. Epub 2014 Jan 30. Antioxid Redox Signal. 2014. PMID: 24180631 Free PMC article.
-
Expression regulation and function of NLRC5.Protein Cell. 2013 Mar;4(3):168-75. doi: 10.1007/s13238-012-2109-3. Epub 2013 Mar 13. Protein Cell. 2013. PMID: 23483478 Free PMC article. Review.
-
Regulation of immune pathways by the NOD-like receptor NLRC5.Immunobiology. 2012 Jan;217(1):13-6. doi: 10.1016/j.imbio.2011.08.011. Epub 2011 Sep 8. Immunobiology. 2012. PMID: 22024701 Free PMC article. Review.
Cited by
-
UHMK1 aids colorectal cancer cell proliferation and chemoresistance through augmenting IL-6/STAT3 signaling.Cell Death Dis. 2022 May 2;13(5):424. doi: 10.1038/s41419-022-04877-8. Cell Death Dis. 2022. PMID: 35501324 Free PMC article.
-
Transcriptional regulation of macrophages in heart failure.Front Cardiovasc Med. 2023 Mar 30;10:1148041. doi: 10.3389/fcvm.2023.1148041. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 37063966 Free PMC article. Review.
-
Adaptor molecules mediate negative regulation of macrophage inflammatory pathways: a closer look.Front Immunol. 2024 Feb 28;15:1355012. doi: 10.3389/fimmu.2024.1355012. eCollection 2024. Front Immunol. 2024. PMID: 38482001 Free PMC article. Review.
-
Macrophage-Specific NLRC5 Protects From Cardiac Remodeling Through Interaction With HSPA8.JACC Basic Transl Sci. 2023 Jan 18;8(5):479-496. doi: 10.1016/j.jacbts.2022.10.001. eCollection 2023 May. JACC Basic Transl Sci. 2023. PMID: 37325412 Free PMC article.
-
Optimized New Shengmai Powder modulation of cAMP/Rap1A signaling pathway attenuates myocardial fibrosis in heart failure.Chin Med. 2024 Feb 24;19(1):30. doi: 10.1186/s13020-024-00902-4. Chin Med. 2024. PMID: 38402401 Free PMC article.
References
-
- Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38:209–23. - PubMed
-
- Akira S, Hemmi H. Recognition of pathogen-associated molecular patterns by TLR family. Immunol Lett. 2003;85:85–95. - PubMed
-
- Sadeghi K, Wessner B, Laggner U, Ploder M, Tamandl D, Friedl J. et al. Vitamin D3 down-regulates monocyte TLR expression and triggers hyporesponsiveness to pathogen-associated molecular patterns. Eur J Immunol. 2006;36:361–70. - PubMed
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

