Viruses in the Nucleus
- PMID: 33753405
- PMCID: PMC8327829
- DOI: 10.1101/cshperspect.a039446
Viruses in the Nucleus
Abstract
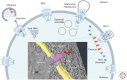
Viral infection is intrinsically linked to the capacity of the virus to generate progeny. Many DNA and some RNA viruses need to access the nuclear machinery and therefore transverse the nuclear envelope barrier through the nuclear pore complex. Viral genomes then become chromatinized either in their episomal form or upon integration into the host genome. Interactions with host DNA, transcription factors or nuclear bodies mediate their replication. Often interfering with nuclear functions, viruses use nuclear architecture to ensure persistent infections. Discovering these multiple modes of replication and persistence served in unraveling many important nuclear processes, such as nuclear trafficking, transcription, and splicing. Here, by using examples of DNA and RNA viral families, we portray the nucleus with the virus inside.
Copyright © 2021 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
Similar articles
-
[Nuclear functions in virus replication].Tanpakushitsu Kakusan Koso. 2006 Nov;51(14 Suppl):2274-86. Tanpakushitsu Kakusan Koso. 2006. PMID: 17471953 Review. Japanese. No abstract available.
-
Nuclear import in viral infections.Curr Top Microbiol Immunol. 2005;285:109-38. doi: 10.1007/3-540-26764-6_4. Curr Top Microbiol Immunol. 2005. PMID: 15609502 Review.
-
Old foes, new understandings: nuclear entry of small non-enveloped DNA viruses.Curr Opin Virol. 2015 Jun;12:59-65. doi: 10.1016/j.coviro.2015.03.017. Epub 2015 Apr 4. Curr Opin Virol. 2015. PMID: 25846849 Review.
-
Nuclear remodelling during viral infections.Cell Microbiol. 2011 Jun;13(6):806-13. doi: 10.1111/j.1462-5822.2011.01596.x. Epub 2011 Apr 28. Cell Microbiol. 2011. PMID: 21501365 Free PMC article. Review.
-
Replication Compartments of DNA Viruses in the Nucleus: Location, Location, Location.Viruses. 2020 Jan 29;12(2):151. doi: 10.3390/v12020151. Viruses. 2020. PMID: 32013091 Free PMC article. Review.
Cited by
-
Targeting the Nucleosome Acidic Patch by Viral Proteins: Two Birds with One Stone?mBio. 2022 Apr 26;13(2):e0173321. doi: 10.1128/mbio.01733-21. Epub 2022 Mar 28. mBio. 2022. PMID: 35343785 Free PMC article. Review.
-
Expanding Insights: Harnessing Expansion Microscopy for Super-Resolution Analysis of HIV-1-Cell Interactions.Viruses. 2024 Oct 15;16(10):1610. doi: 10.3390/v16101610. Viruses. 2024. PMID: 39459943 Free PMC article.
-
Interplay between Lipid Metabolism, Lipid Droplets, and DNA Virus Infections.Cells. 2022 Jul 17;11(14):2224. doi: 10.3390/cells11142224. Cells. 2022. PMID: 35883666 Free PMC article. Review.
-
Contribution of carbohydrate-related metabolism in Herpesvirus infections.Curr Res Microb Sci. 2023 May 17;4:100192. doi: 10.1016/j.crmicr.2023.100192. eCollection 2023. Curr Res Microb Sci. 2023. PMID: 37273578 Free PMC article.
-
Chromatin accessibility: methods, mechanisms, and biological insights.Nucleus. 2022 Dec;13(1):236-276. doi: 10.1080/19491034.2022.2143106. Nucleus. 2022. PMID: 36404679 Free PMC article. Review.
References
-
- Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, et al. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci 107: 5563–5568. 10.1073/pnas.0913586107 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
