CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing
- PMID: 33750389
- PMCID: PMC7941935
- DOI: 10.1186/s12943-021-01333-7
CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing
Erratum in
-
Correction: CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-IRNA-editing.Mol Cancer. 2022 Aug 19;21(1):165. doi: 10.1186/s12943-022-01636-3. Mol Cancer. 2022. PMID: 35986360 Free PMC article. No abstract available.
Abstract
Background: A growing number of studies have focused on investigating circRNAs as crucial regulators in the progression of multiple cancer types. Nevertheless, the biological effects and underlying mechanisms of circRNAs in pancreatic ductal adenocarcinoma (PDAC) remain unclear.
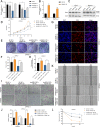
Methods: Differentially expressed circRNAs between cancerous tissue and adjacent normal tissues were identified by RNA sequencing in PDAC. Subsequently, in vitro and in vivo functional experiments were performed to investigate the functional roles of circNEIL3 in PDAC tumour growth and metastasis. Furthermore, RNA pull-down, dual-luciferase reporter assays, RNA immunoprecipitation (RIP) assays, fluorescent in situ hybridization (FISH) and Sanger sequencing assays were performed to examine the circular interaction among circNEIL3, miR-432-5p and adenosine deaminases acting on RNA 1 (ADAR1).
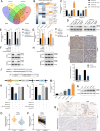
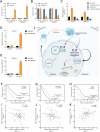
Results: CircNEIL3 was upregulated in PDAC and promoted the progression of PDAC cells both in vitro and in vivo. Mechanistically, circNEIL3 was shown to regulate the expression of ADAR1 by sponging miR-432-5p to induce RNA editing of glioma-associated oncogene 1 (GLI1), ultimately influencing cell cycle progression and promoting epithelial-to-mesenchymal transition (EMT) in PDAC cells. Moreover, we discovered that the circNEIL3/miR-432-5p/ADAR1 axis was correlated with the PDAC clinical stage and overall survival of PDAC patients, while ADAR1 may reduce the biogenesis of circNEIL3.
Conclusions: Our findings reveal that circNEIL3 facilitates the proliferation and metastasis of PDAC through the circNEIL3/miR-432-5p/ADAR1/GLI1/cell cycle and EMT axis and that its expression is regulated by ADAR1 through a negative feedback loop. Therefore, circNEIL3 may serve as a prognostic marker and a therapeutic target for PDAC.
Keywords: ADAR1; Cell cycle; EMT; GLI1; Pancreatic ductal adenocarcinoma; RNA editing; circNEIL3; miR-432-5p.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma.J Exp Clin Cancer Res. 2021 Aug 20;40(1):261. doi: 10.1186/s13046-021-02063-w. J Exp Clin Cancer Res. 2021. PMID: 34416910 Free PMC article.
-
CircPTPRA promotes the progression of pancreatic ductal adenocarcinoma via the miR-140-5p/LMNB1 axis.Cancer Med. 2023 May;12(10):11651-11671. doi: 10.1002/cam4.5869. Epub 2023 Apr 11. Cancer Med. 2023. PMID: 37041721 Free PMC article.
-
Circular RNA circBFAR promotes the progression of pancreatic ductal adenocarcinoma via the miR-34b-5p/MET/Akt axis.Mol Cancer. 2020 May 6;19(1):83. doi: 10.1186/s12943-020-01196-4. Mol Cancer. 2020. PMID: 32375768 Free PMC article.
-
The involvement of lncRNAs in the development and progression of pancreatic cancer.Cancer Biol Ther. 2017 Dec 2;18(12):927-936. doi: 10.1080/15384047.2017.1385682. Epub 2017 Nov 27. Cancer Biol Ther. 2017. PMID: 29053398 Free PMC article. Review.
-
MicroRNAs as Indicators of Malignancy in Pancreatic Ductal Adenocarcinoma (PDAC) and Cystic Pancreatic Lesions.Cells. 2022 Aug 2;11(15):2374. doi: 10.3390/cells11152374. Cells. 2022. PMID: 35954223 Free PMC article. Review.
Cited by
-
Autophagy-Related ncRNAs in Pancreatic Cancer.Pharmaceuticals (Basel). 2022 Dec 13;15(12):1547. doi: 10.3390/ph15121547. Pharmaceuticals (Basel). 2022. PMID: 36558998 Free PMC article. Review.
-
Targeting miRNA by CRISPR/Cas in cancer: advantages and challenges.Mil Med Res. 2023 Jul 17;10(1):32. doi: 10.1186/s40779-023-00468-6. Mil Med Res. 2023. PMID: 37460924 Free PMC article. Review.
-
A-to-I RNA editing by ADAR and its therapeutic applications: From viral infections to cancer immunotherapy.Wiley Interdiscip Rev RNA. 2023 Sep 17:e1817. doi: 10.1002/wrna.1817. Online ahead of print. Wiley Interdiscip Rev RNA. 2023. PMID: 37718249 Review.
-
N6-methyladenosine-modified circIGF2BP3 inhibits CD8+ T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer.Mol Cancer. 2021 Aug 20;20(1):105. doi: 10.1186/s12943-021-01398-4. Mol Cancer. 2021. PMID: 34416901 Free PMC article.
-
circPTPN22 attenuates immune microenvironment of pancreatic cancer via STAT3 acetylation.Cancer Gene Ther. 2023 Apr;30(4):559-566. doi: 10.1038/s41417-021-00382-w. Epub 2021 Sep 1. Cancer Gene Ther. 2023. PMID: 34471233
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

