Precision Surface Microtopography Regulates Cell Fate via Changes to Actomyosin Contractility and Nuclear Architecture
- PMID: 33747730
- PMCID: PMC7967085
- DOI: 10.1002/advs.202003186
Precision Surface Microtopography Regulates Cell Fate via Changes to Actomyosin Contractility and Nuclear Architecture
Abstract
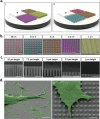
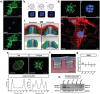
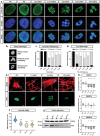
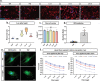
Cells are able to perceive complex mechanical cues from their microenvironment, which in turn influences their development. Although the understanding of these intricate mechanotransductive signals is evolving, the precise roles of substrate microtopography in directing cell fate is still poorly understood. Here, UV nanoimprint lithography is used to generate micropillar arrays ranging from 1 to 10 µm in height, width, and spacing to investigate the impact of microtopography on mechanotransduction. Using mesenchymal stem cells (MSCs) as a model, stark pattern-specific changes in nuclear architecture, lamin A/C accumulation, chromatin positioning, and DNA methyltransferase expression, are demonstrated. MSC osteogenesis is also enhanced specifically on micropillars with 5 µm width/spacing and 5 µm height. Intriguingly, the highest degree of osteogenesis correlates with patterns that stimulated maximal nuclear deformation which is shown to be dependent on myosin-II-generated tension. The outcomes determine new insights into nuclear mechanotransduction by demonstrating that force transmission across the nuclear envelope can be modulated by substrate topography, and that this can alter chromatin organisation and impact upon cell fate. These findings have potential to inform the development of microstructured cell culture substrates that can direct cell mechanotransduction and fate for therapeutic applications in both research and clinical sectors.
Keywords: mechanotransduction; mesenchymal stem/stromal cells; microtopography; osteogenesis.
© 2021 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Early committed polarization of intracellular tension in response to cell shape determines the osteogenic differentiation of mesenchymal stromal cells.Acta Biomater. 2023 Jun;163:287-301. doi: 10.1016/j.actbio.2022.10.052. Epub 2022 Nov 1. Acta Biomater. 2023. PMID: 36328121 Free PMC article.
-
Microtopography-Induced Nuclear Deformation Triggers Chromatin Reorganization and Cytoskeleton Remodeling.Chem Biomed Imaging. 2024 May 10;2(7):481-489. doi: 10.1021/cbmi.4c00035. eCollection 2024 Jul 22. Chem Biomed Imaging. 2024. PMID: 39473534 Free PMC article.
-
Integration of Mesenchymal Stem Cells into a Novel Micropillar Confinement Assay.Tissue Eng Part C Methods. 2019 Nov;25(11):662-676. doi: 10.1089/ten.TEC.2019.0083. Epub 2019 Sep 11. Tissue Eng Part C Methods. 2019. PMID: 31347455 Free PMC article.
-
Mechanobiology of mesenchymal stem cells: Perspective into mechanical induction of MSC fate.Acta Biomater. 2015 Jul;20:1-9. doi: 10.1016/j.actbio.2015.04.008. Epub 2015 Apr 11. Acta Biomater. 2015. PMID: 25871537 Review.
-
Mechanical regulation of mesenchymal stem cell differentiation.J Anat. 2015 Dec;227(6):717-31. doi: 10.1111/joa.12243. Epub 2014 Nov 9. J Anat. 2015. PMID: 25382217 Free PMC article. Review.
Cited by
-
Extracellular Matrix Cues Regulate Mechanosensing and Mechanotransduction of Cancer Cells.Cells. 2024 Jan 2;13(1):96. doi: 10.3390/cells13010096. Cells. 2024. PMID: 38201302 Free PMC article. Review.
-
Chromatin reprogramming and bone regeneration in vitro and in vivo via the microtopography-induced constriction of cell nuclei.Nat Biomed Eng. 2023 Nov;7(11):1514-1529. doi: 10.1038/s41551-023-01053-x. Epub 2023 Jun 12. Nat Biomed Eng. 2023. PMID: 37308586 Free PMC article.
-
Effect of Functionalization of Texturized Polypropylene Surface by Silanization and HBII-RGD Attachment on Response of Primary Abdominal and Vaginal Fibroblasts.Polymers (Basel). 2024 Feb 29;16(5):667. doi: 10.3390/polym16050667. Polymers (Basel). 2024. PMID: 38475352 Free PMC article.
-
Advances in lithographic techniques for precision nanostructure fabrication in biomedical applications.Discov Nano. 2023 Dec 11;18(1):153. doi: 10.1186/s11671-023-03938-x. Discov Nano. 2023. PMID: 38082047 Free PMC article. Review.
-
Integrating physicomechanical and biological strategies for BTE: biomaterials-induced osteogenic differentiation of MSCs.Theranostics. 2023 May 21;13(10):3245-3275. doi: 10.7150/thno.84759. eCollection 2023. Theranostics. 2023. PMID: 37351163 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
