Cooperation between oncogenic Ras and wild-type p53 stimulates STAT non-cell autonomously to promote tumor radioresistance
- PMID: 33742110
- PMCID: PMC7979758
- DOI: 10.1038/s42003-021-01898-5
Cooperation between oncogenic Ras and wild-type p53 stimulates STAT non-cell autonomously to promote tumor radioresistance
Abstract
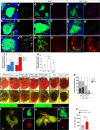
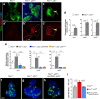
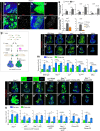
Oncogenic RAS mutations are associated with tumor resistance to radiation therapy. Cell-cell interactions in the tumor microenvironment (TME) profoundly influence therapy outcomes. However, the nature of these interactions and their role in Ras tumor radioresistance remain unclear. Here we use Drosophila oncogenic Ras tissues and human Ras cancer cell radiation models to address these questions. We discover that cellular response to genotoxic stress cooperates with oncogenic Ras to activate JAK/STAT non-cell autonomously in the TME. Specifically, p53 is heterogeneously activated in Ras tumor tissues in response to irradiation. This mosaicism allows high p53-expressing Ras clones to stimulate JAK/STAT cytokines, which activate JAK/STAT in the nearby low p53-expressing surviving Ras clones, leading to robust tumor re-establishment. Blocking any part of this cell-cell communication loop re-sensitizes Ras tumor cells to irradiation. These findings suggest that coupling STAT inhibitors to radiotherapy might improve clinical outcomes for Ras cancer patients.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion.Nature. 2010 Jan 28;463(7280):545-8. doi: 10.1038/nature08702. Epub 2010 Jan 13. Nature. 2010. PMID: 20072127 Free PMC article.
-
Gα73B is a downstream effector of JAK/STAT signalling and a regulator of Rho1 in Drosophila haematopoiesis.J Cell Sci. 2014 Jan 1;127(Pt 1):101-10. doi: 10.1242/jcs.132852. Epub 2013 Oct 25. J Cell Sci. 2014. PMID: 24163435
-
GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo.Gene Expr Patterns. 2007 Jan;7(3):323-31. doi: 10.1016/j.modgep.2006.08.003. Epub 2006 Aug 22. Gene Expr Patterns. 2007. PMID: 17008134
-
Organogenesis and tumorigenesis: insight from the JAK/STAT pathway in the Drosophila eye.Dev Dyn. 2010 Oct;239(10):2522-33. doi: 10.1002/dvdy.22394. Dev Dyn. 2010. PMID: 20737505 Free PMC article. Review.
-
JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions.Development. 2006 Jul;133(14):2605-16. doi: 10.1242/dev.02411. Development. 2006. PMID: 16794031 Review.
Cited by
-
Lipid Metabolism Regulates Oxidative Stress and Ferroptosis in RAS-Driven Cancers: A Perspective on Cancer Progression and Therapy.Front Mol Biosci. 2021 Aug 16;8:706650. doi: 10.3389/fmolb.2021.706650. eCollection 2021. Front Mol Biosci. 2021. PMID: 34485382 Free PMC article. Review.
-
Tumor suppressor p53 mediates interleukin-6 expression to enable cancer cell evasion of genotoxic stress.Cell Death Discov. 2023 Sep 11;9(1):340. doi: 10.1038/s41420-023-01638-0. Cell Death Discov. 2023. PMID: 37696858 Free PMC article.
-
Mechanisms of Germline Stem Cell Competition across Species.Life (Basel). 2024 Oct 1;14(10):1251. doi: 10.3390/life14101251. Life (Basel). 2024. PMID: 39459551 Free PMC article. Review.
-
p53 Signaling on Microenvironment and Its Contribution to Tissue Chemoresistance.Membranes (Basel). 2022 Feb 9;12(2):202. doi: 10.3390/membranes12020202. Membranes (Basel). 2022. PMID: 35207121 Free PMC article. Review.
-
DNA repair in tumor radioresistance: insights from fruit flies genetics.Cell Oncol (Dordr). 2024 Jun;47(3):717-732. doi: 10.1007/s13402-023-00906-6. Epub 2023 Dec 14. Cell Oncol (Dordr). 2024. PMID: 38095764 Review.
References
-
- Bernhard EJ, et al. Direct evidence for the contribution of activated N-ras and K-ras oncogenes to increased intrinsic radiation resistance in human tumor cell lines. Cancer Res. 2000;60:6597–6600. - PubMed
-
- Gupta AK, et al. The Ras radiation resistance pathway. Cancer Res. 2001;61:4278–4282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

