Negative elongation factor regulates muscle progenitor expansion for efficient myofiber repair and stem cell pool repopulation
- PMID: 33735618
- PMCID: PMC8357161
- DOI: 10.1016/j.devcel.2021.02.025
Negative elongation factor regulates muscle progenitor expansion for efficient myofiber repair and stem cell pool repopulation
Abstract
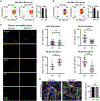
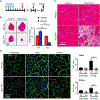
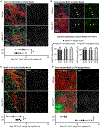
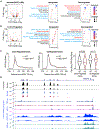
Negative elongation factor (NELF) is a critical transcriptional regulator that stabilizes paused RNA polymerase to permit rapid gene expression changes in response to environmental cues. Although NELF is essential for embryonic development, its role in adult stem cells remains unclear. In this study, through a muscle-stem-cell-specific deletion, we showed that NELF is required for efficient muscle regeneration and stem cell pool replenishment. In mechanistic studies using PRO-seq, single-cell trajectory analyses and myofiber cultures revealed that NELF works at a specific stage of regeneration whereby it modulates p53 signaling to permit massive expansion of muscle progenitors. Strikingly, transplantation experiments indicated that these progenitors are also necessary for stem cell pool repopulation, implying that they are able to return to quiescence. Thus, we identified a critical role for NELF in the expansion of muscle progenitors in response to injury and revealed that progenitors returning to quiescence are major contributors to the stem cell pool repopulation.
Keywords: NELF; PEDF signaling; muscle regeneration; muscle stem cells; nascent transcript stability; p53 signaling; promoter proximal pausing; stem cell niche; stem cell self-renewal; transcriptional regulation.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Similar articles
-
Zfp423 Regulates Skeletal Muscle Regeneration and Proliferation.Mol Cell Biol. 2019 Apr 2;39(8):e00447-18. doi: 10.1128/MCB.00447-18. Print 2019 Apr 15. Mol Cell Biol. 2019. PMID: 30692273 Free PMC article.
-
Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration.J Cell Biol. 2012 Sep 3;198(5):815-32. doi: 10.1083/jcb.201201050. J Cell Biol. 2012. PMID: 22945933 Free PMC article.
-
Single EDL Myofiber Isolation for Analyses of Quiescent and Activated Muscle Stem Cells.Methods Mol Biol. 2018;1686:149-159. doi: 10.1007/978-1-4939-7371-2_11. Methods Mol Biol. 2018. PMID: 29030819
-
Niche regulation of muscle satellite cell self-renewal and differentiation.Cell Stem Cell. 2008 Jan 10;2(1):22-31. doi: 10.1016/j.stem.2007.12.012. Cell Stem Cell. 2008. PMID: 18371418 Review.
-
Intrinsic and extrinsic mechanisms regulating satellite cell function.Development. 2015 May 1;142(9):1572-81. doi: 10.1242/dev.114223. Development. 2015. PMID: 25922523 Free PMC article. Review.
Cited by
-
Chromatin and transcription factor profiling in rare stem cell populations using CUT&Tag.STAR Protoc. 2021 Aug 19;2(3):100751. doi: 10.1016/j.xpro.2021.100751. eCollection 2021 Sep 17. STAR Protoc. 2021. PMID: 34467227 Free PMC article.
-
Dynamic RNA polymerase II occupancy drives differentiation of the intestine under the direction of HNF4.Cell Rep. 2024 Jun 25;43(6):114242. doi: 10.1016/j.celrep.2024.114242. Epub 2024 May 19. Cell Rep. 2024. PMID: 38768033 Free PMC article.
-
The mini-IDLE 3D biomimetic culture assay enables interrogation of mechanisms governing muscle stem cell quiescence and niche repopulation.Elife. 2022 Dec 20;11:e81738. doi: 10.7554/eLife.81738. Elife. 2022. PMID: 36537758 Free PMC article.
-
Beyond the bulk: overview and novel insights into the dynamics of muscle satellite cells during muscle regeneration.Inflamm Regen. 2024 Sep 26;44(1):39. doi: 10.1186/s41232-024-00354-1. Inflamm Regen. 2024. PMID: 39327631 Free PMC article. Review.
-
RNA polymerase II pausing factor NELF in CD8+ T cells promotes antitumor immunity.Nat Commun. 2022 Apr 20;13(1):2155. doi: 10.1038/s41467-022-29869-2. Nat Commun. 2022. PMID: 35444206 Free PMC article.
References
-
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, and Lis JT (2005). Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17, 103–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

