Genomic regions associated with microdeletion/microduplication syndromes exhibit extreme diversity of structural variation
- PMID: 33724415
- PMCID: PMC8045732
- DOI: 10.1093/genetics/iyaa038
Genomic regions associated with microdeletion/microduplication syndromes exhibit extreme diversity of structural variation
Abstract
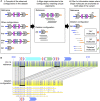
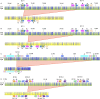
Segmental duplications (SDs) are a class of long, repetitive DNA elements whose paralogs share a high level of sequence similarity with each other. SDs mediate chromosomal rearrangements that lead to structural variation in the general population as well as genomic disorders associated with multiple congenital anomalies, including the 7q11.23 (Williams-Beuren Syndrome, WBS), 15q13.3, and 16p12.2 microdeletion syndromes. Population-level characterization of SDs has generally been lacking because most techniques used for analyzing these complex regions are both labor and cost intensive. In this study, we have used a high-throughput technique to genotype complex structural variation with a single molecule, long-range optical mapping approach. We characterized SDs and identified novel structural variants (SVs) at 7q11.23, 15q13.3, and 16p12.2 using optical mapping data from 154 phenotypically normal individuals from 26 populations comprising five super-populations. We detected several novel SVs for each locus, some of which had significantly different prevalence between populations. Additionally, we localized the microdeletion breakpoints to specific paralogous duplicons located within complex SDs in two patients with WBS, one patient with 15q13.3, and one patient with 16p12.2 microdeletion syndromes. The population-level data presented here highlights the extreme diversity of large and complex SVs within SD-containing regions. The approach we outline will greatly facilitate the investigation of the role of inter-SD structural variation as a driver of chromosomal rearrangements and genomic disorders.
Keywords: genome mapping; genomic disorders; segmental duplications; structural variation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



Similar articles
-
Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability.Nat Genet. 2014 Dec;46(12):1293-302. doi: 10.1038/ng.3120. Epub 2014 Oct 19. Nat Genet. 2014. PMID: 25326701 Free PMC article.
-
Clinical utility gene card for: 16p12.2 microdeletion.Eur J Hum Genet. 2017 Feb;25(2). doi: 10.1038/ejhg.2016.158. Epub 2016 Nov 16. Eur J Hum Genet. 2017. PMID: 27848943 Free PMC article. No abstract available.
-
Genome rearrangements detected by SNP microarrays in individuals with intellectual disability referred with possible Williams syndrome.PLoS One. 2010 Aug 31;5(8):e12349. doi: 10.1371/journal.pone.0012349. PLoS One. 2010. PMID: 20824207 Free PMC article.
-
An Update on Common Chromosome Microdeletion and Microduplication Syndromes.Pediatr Ann. 2018 May 1;47(5):e198-e203. doi: 10.3928/19382359-20180419-01. Pediatr Ann. 2018. PMID: 29750287 Review.
-
Compound heterozygous microdeletion of chromosome 15q13.3 region in a child with hypotonia, impaired vision, and global developmental delay.Am J Med Genet A. 2014 Jul;164A(7):1815-20. doi: 10.1002/ajmg.a.36535. Epub 2014 Apr 3. Am J Med Genet A. 2014. PMID: 24700535 Review.
Cited by
-
High level of complexity and global diversity of the 3q29 locus revealed by optical mapping and long-read sequencing.Genome Med. 2023 May 10;15(1):35. doi: 10.1186/s13073-023-01184-5. Genome Med. 2023. PMID: 37165454 Free PMC article.
-
Optical genome mapping in acute myeloid leukemia: a multicenter evaluation.Blood Adv. 2023 Apr 11;7(7):1297-1307. doi: 10.1182/bloodadvances.2022007583. Blood Adv. 2023. PMID: 36417763 Free PMC article.
-
Impact and characterization of serial structural variations across humans and great apes.Nat Commun. 2024 Sep 13;15(1):8007. doi: 10.1038/s41467-024-52027-9. Nat Commun. 2024. PMID: 39266513 Free PMC article.
-
Comparative Benchmarking of Optical Genome Mapping and Chromosomal Microarray Reveals High Technological Concordance in CNV Identification and Additional Structural Variant Refinement.Genes (Basel). 2023 Sep 26;14(10):1868. doi: 10.3390/genes14101868. Genes (Basel). 2023. PMID: 37895217 Free PMC article.
-
Optical Genome Mapping as a Potential Routine Clinical Diagnostic Method.Genes (Basel). 2024 Mar 7;15(3):342. doi: 10.3390/genes15030342. Genes (Basel). 2024. PMID: 38540401 Free PMC article.
References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

