Impaired phosphatidylethanolamine metabolism activates a reversible stress response that detects and resolves mutant mitochondrial precursors
- PMID: 33718843
- PMCID: PMC7921845
- DOI: 10.1016/j.isci.2021.102196
Impaired phosphatidylethanolamine metabolism activates a reversible stress response that detects and resolves mutant mitochondrial precursors
Abstract
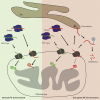
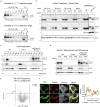
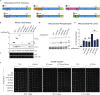
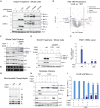
Phosphatidylethanolamine (PE) made in mitochondria has long been recognized as an important precursor for phosphatidylcholine production that occurs in the endoplasmic reticulum (ER). Recently, the strict mitochondrial localization of the enzyme that makes PE in the mitochondrion, phosphatidylserine decarboxylase 1 (Psd1), was questioned. Since a dual localization of Psd1 to the ER would have far-reaching implications, we initiated our study to independently re-assess the subcellular distribution of Psd1. Our results support the unavoidable conclusion that the vast majority, if not all, of functional Psd1 resides in the mitochondrion. Through our efforts, we discovered that mutant forms of Psd1 that impair a self-processing step needed for it to become functional are dually localized to the ER when expressed in a PE-limiting environment. We conclude that severely impaired cellular PE metabolism provokes an ER-assisted adaptive response that is capable of identifying and resolving nonfunctional mitochondrial precursors.
Keywords: Cell Biology; Molecular Physiology; Proteomics.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Lipid Homeostasis Is Maintained by Dual Targeting of the Mitochondrial PE Biosynthesis Enzyme to the ER.Dev Cell. 2018 Jan 22;44(2):261-270.e6. doi: 10.1016/j.devcel.2017.11.023. Epub 2017 Dec 28. Dev Cell. 2018. PMID: 29290583 Free PMC article.
-
Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1.J Biol Chem. 2012 Oct 26;287(44):36744-55. doi: 10.1074/jbc.M112.398107. Epub 2012 Sep 13. J Biol Chem. 2012. PMID: 22984266 Free PMC article.
-
Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria.Plant Physiol. 2007 Jun;144(2):904-14. doi: 10.1104/pp.107.095414. Epub 2007 Apr 20. Plant Physiol. 2007. PMID: 17449644 Free PMC article.
-
Molecular and cell biology of phosphatidylserine and phosphatidylethanolamine metabolism.Prog Nucleic Acid Res Mol Biol. 2003;75:69-111. doi: 10.1016/s0079-6603(03)75003-x. Prog Nucleic Acid Res Mol Biol. 2003. PMID: 14604010 Review.
-
Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells.Biochim Biophys Acta. 2013 Mar;1831(3):543-54. doi: 10.1016/j.bbalip.2012.08.016. Epub 2012 Aug 29. Biochim Biophys Acta. 2013. PMID: 22960354 Review.
Cited by
-
Chemical inhibition of phosphatidylcholine biogenesis reveals its role in mitochondrial division.iScience. 2024 Feb 10;27(3):109189. doi: 10.1016/j.isci.2024.109189. eCollection 2024 Mar 15. iScience. 2024. PMID: 38420588 Free PMC article.
-
Dissociation of ERMES clusters plays a key role in attenuating the endoplasmic reticulum stress.iScience. 2022 Oct 14;25(11):105362. doi: 10.1016/j.isci.2022.105362. eCollection 2022 Nov 18. iScience. 2022. PMID: 36339260 Free PMC article.
-
ER-localized phosphatidylethanolamine synthase plays a conserved role in lipid droplet formation.Mol Biol Cell. 2022 Jan 1;33(1):ar11. doi: 10.1091/mbc.E21-11-0558-T. Epub 2021 Nov 24. Mol Biol Cell. 2022. PMID: 34818062 Free PMC article.
-
The emergent role of mitochondrial surveillance in cellular health.Aging Cell. 2022 Nov;21(11):e13710. doi: 10.1111/acel.13710. Epub 2022 Sep 11. Aging Cell. 2022. PMID: 36088658 Free PMC article. Review.
-
Mitochondrial compartmentalization: emerging themes in structure and function.Trends Biochem Sci. 2021 Nov;46(11):902-917. doi: 10.1016/j.tibs.2021.06.003. Epub 2021 Jul 7. Trends Biochem Sci. 2021. PMID: 34244035 Free PMC article. Review.
References
-
- Achleitner G., Zweytick D., Trotter P.J., Voelker D.R., Daum G. Synthesis and intracellular transport of aminoglycerophospholipids in permeabilized cells of the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:29836–29842. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

