Detection and Genetic Diversity of Porcine Coronavirus Involved in Diarrhea Outbreaks in Spain
- PMID: 33718476
- PMCID: PMC7947225
- DOI: 10.3389/fvets.2021.651999
Detection and Genetic Diversity of Porcine Coronavirus Involved in Diarrhea Outbreaks in Spain
Abstract
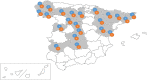
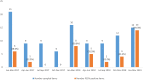
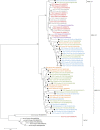
Porcine enteric coronaviruses include some of the most relevant viral pathogens to the swine industry such as porcine epidemic diarrhea virus (PEDV) or porcine transmissible gastroenteritis virus (TGEV) as well as several recently identified virus such as swine enteric coronavirus (SeCoV), porcine deltacoronavirus (PDCoV) or swine enteric alphacoronavirus (SeACoV). The aim of this study is the identification and characterization of enteric coronaviruses on Spanish pig farms between 2017 and 2019. The study was carried out on 106 swine farms with diarrhea outbreaks where a viral etiology was suspected by using two duplex RT-PCRs developed for the detection of porcine enteric coronaviruses. PEDV was the only coronavirus detected in our research (38.7% positive outbreaks, 41 out of 106) and neither TGEV, SeCoV, PDCoV nor SeACoV were detected in any of the samples. The complete S-gene of all the PEDV isolates recovered were obtained and compared to PEDV and SeCoV sequences available in GenBank. The phylogenetic tree showed that only PEDV of the INDEL 2 or G1b genogroup has circulated in Spain between 2017 and 2019. Three different variants were detected, the recombinant PEDV-SeCoV being the most widespread. These results show that PEDV is a relevant cause of enteric disorders in pigs in Spain while new emerging coronavirus have not been detected so far. However, the monitoring of these virus is advisable to curtail their emergence and spread.
Keywords: INDEL strain; PEDV; S or Spike gene; pig; swine coronavirus.
Copyright © 2021 Puente, Argüello, Mencía-Ares, Gómez-García, Rubio and Carvajal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
A retrospective study of porcine epidemic diarrhoea virus (PEDV) reveals the presence of swine enteric coronavirus (SeCoV) since 1993 and the recent introduction of a recombinant PEDV-SeCoV in Spain.Transbound Emerg Dis. 2020 Nov;67(6):2911-2922. doi: 10.1111/tbed.13666. Epub 2020 Jul 1. Transbound Emerg Dis. 2020. PMID: 32511876
-
Multiplex Real-Time RT-PCR Assays for Detection and Differentiation of Porcine Enteric Coronaviruses.Pathogens. 2023 Aug 14;12(8):1040. doi: 10.3390/pathogens12081040. Pathogens. 2023. PMID: 37624000 Free PMC article.
-
Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019.PLoS One. 2021 Jun 24;16(6):e0253622. doi: 10.1371/journal.pone.0253622. eCollection 2021. PLoS One. 2021. PMID: 34166425 Free PMC article.
-
Swine Enteric Coronavirus: Diverse Pathogen-Host Interactions.Int J Mol Sci. 2022 Apr 2;23(7):3953. doi: 10.3390/ijms23073953. Int J Mol Sci. 2022. PMID: 35409315 Free PMC article. Review.
-
Interplay between swine enteric coronaviruses and host innate immune.Front Vet Sci. 2022 Dec 22;9:1083605. doi: 10.3389/fvets.2022.1083605. eCollection 2022. Front Vet Sci. 2022. PMID: 36619958 Free PMC article. Review.
Cited by
-
Death Receptor DR5 as a Proviral Factor for Viral Entry and Replication of Coronavirus PEDV.Viruses. 2022 Dec 6;14(12):2724. doi: 10.3390/v14122724. Viruses. 2022. PMID: 36560727 Free PMC article.
-
Occurrence of Rotavirus A Genotypes and Other Enteric Pathogens in Diarrheic Suckling Piglets from Spanish Swine Farms.Animals (Basel). 2022 Jan 20;12(3):251. doi: 10.3390/ani12030251. Animals (Basel). 2022. PMID: 35158575 Free PMC article.
-
Development of a multiplex reverse transcription-quantitative PCR (qPCR) method for detecting common causative agents of swine viral diarrhea in China.Porcine Health Manag. 2024 Mar 5;10(1):12. doi: 10.1186/s40813-024-00364-y. Porcine Health Manag. 2024. PMID: 38444040 Free PMC article.
-
Novel Neutralizing Epitope of PEDV S1 Protein Identified by IgM Monoclonal Antibody.Viruses. 2022 Jan 11;14(1):125. doi: 10.3390/v14010125. Viruses. 2022. PMID: 35062329 Free PMC article.
-
The impact and complete genome characterisation of viruses involved in outbreaks of gastroenteritis in a farrow-to-finish holding.Sci Rep. 2023 Oct 31;13(1):18780. doi: 10.1038/s41598-023-45994-4. Sci Rep. 2023. PMID: 37907693 Free PMC article.
References
-
- Saif LJ, Wang Q, Vlasova AN, Jung K, Xiao S. Coronaviruses. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, Zhang J. editors. Diseases of Swine. Hoboken: John Wiley and Sons. (2019) p. 488−523. 10.1002/9781119350927.ch31 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

