The p97-UBXN1 complex regulates aggresome formation
- PMID: 33712450
- PMCID: PMC8077447
- DOI: 10.1242/jcs.254201
The p97-UBXN1 complex regulates aggresome formation
Abstract
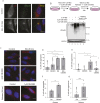
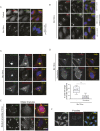
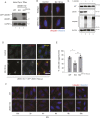
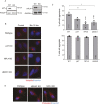
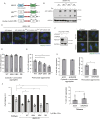
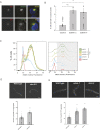
The recognition and disposal of misfolded proteins is essential for the maintenance of cellular homeostasis. Perturbations in the pathways that promote degradation of aberrant proteins contribute to a variety of protein aggregation disorders broadly termed proteinopathies. The AAA-ATPase p97 (also known as VCP), in combination with adaptor proteins, functions to identify ubiquitylated proteins and target them for degradation by the proteasome or through autophagy. Mutations in p97 cause multi-system proteinopathies; however, the precise defects underlying these disorders are unclear. Here, we systematically investigate the role of p97 and its adaptors in the process of formation of aggresomes, membrane-less structures containing ubiquitylated proteins that arise upon proteasome inhibition. We demonstrate that p97 mediates aggresome formation and clearance, and identify a novel role for the adaptor UBXN1 in the process of aggresome formation. UBXN1 is recruited to aggresomes, and UBXN1-knockout cells are unable to form aggresomes. Loss of p97-UBXN1 results in increased Huntingtin polyQ inclusion bodies both in mammalian cells and in a C. elegans model of Huntington's disease. Together, our results identify evolutionarily conserved roles for p97-UBXN1 in the disposal of protein aggregates.
Keywords: Aggregate; Aggresome; Inclusion body; PolyQ; Proteasome; Ubiquitin.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
Similar articles
-
The VCP-UBXN1 Complex Mediates Triage of Ubiquitylated Cytosolic Proteins Bound to the BAG6 Complex.Mol Cell Biol. 2018 Jun 14;38(13):e00154-18. doi: 10.1128/MCB.00154-18. Print 2018 Jul 1. Mol Cell Biol. 2018. PMID: 29685906 Free PMC article.
-
VCP/p97 cofactor UBXN1/SAKS1 regulates mitophagy by modulating MFN2 removal from mitochondria.Autophagy. 2022 Jan;18(1):171-190. doi: 10.1080/15548627.2021.1922982. Epub 2021 May 9. Autophagy. 2022. PMID: 33966597 Free PMC article.
-
Valosin-containing Protein (VCP)/p97 Segregase Mediates Proteolytic Processing of Cockayne Syndrome Group B (CSB) in Damaged Chromatin.J Biol Chem. 2016 Apr 1;291(14):7396-408. doi: 10.1074/jbc.M115.705350. Epub 2016 Jan 29. J Biol Chem. 2016. PMID: 26826127 Free PMC article.
-
Insights into adaptor binding to the AAA protein p97.Biochem Soc Trans. 2008 Feb;36(Pt 1):62-7. doi: 10.1042/BST0360062. Biochem Soc Trans. 2008. PMID: 18208387 Review.
-
Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system.Nat Cell Biol. 2012 Feb 2;14(2):117-23. doi: 10.1038/ncb2407. Nat Cell Biol. 2012. PMID: 22298039 Review.
Cited by
-
Interactome screening of C9orf72 dipeptide repeats reveals VCP sequestration and functional impairment by polyGA.Brain. 2022 Apr 18;145(2):684-699. doi: 10.1093/brain/awab300. Brain. 2022. PMID: 34534264 Free PMC article.
-
Role of ubiquitin regulatory X domain‑containing protein 3B in the development of hepatocellular carcinoma (Review).Oncol Rep. 2023 Mar;49(3):57. doi: 10.3892/or.2023.8494. Epub 2023 Feb 17. Oncol Rep. 2023. PMID: 36799187 Free PMC article. Review.
-
Precision Proteoform Design for 4R Tau Isoform Selective Templated Aggregation.bioRxiv [Preprint]. 2023 Nov 28:2023.08.31.555649. doi: 10.1101/2023.08.31.555649. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Apr 9;121(15):e2320456121. doi: 10.1073/pnas.2320456121. PMID: 37693456 Free PMC article. Updated. Preprint.
-
Intermediate filaments associate with aggresome-like structures in proteostressed C. elegans neurons and influence large vesicle extrusions as exophers.Nat Commun. 2023 Jul 24;14(1):4450. doi: 10.1038/s41467-023-39700-1. Nat Commun. 2023. PMID: 37488107 Free PMC article.
-
Characterization of Skeletal Muscle Biopsy and Derived Myoblasts in a Patient Carrying Arg14del Mutation in Phospholamban Gene.Cells. 2023 May 17;12(10):1405. doi: 10.3390/cells12101405. Cells. 2023. PMID: 37408239 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

