Comparative transcriptomic analysis of thermally stressed Arabidopsis thaliana meiotic recombination mutants
- PMID: 33711924
- PMCID: PMC7953577
- DOI: 10.1186/s12864-021-07497-2
Comparative transcriptomic analysis of thermally stressed Arabidopsis thaliana meiotic recombination mutants
Abstract
Background: Meiosis is a specialized cell division that underpins sexual reproduction in most eukaryotes. During meiosis, interhomolog meiotic recombination facilitates accurate chromosome segregation and generates genetic diversity by shuffling parental alleles in the gametes. The frequency of meiotic recombination in Arabidopsis has a U-shaped curve in response to environmental temperature, and is dependent on the Type I, crossover (CO) interference-sensitive pathway. The mechanisms that modulate recombination frequency in response to temperature are not yet known.
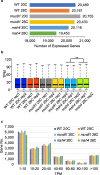
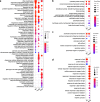
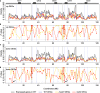
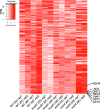
Results: In this study, we compare the transcriptomes of thermally-stressed meiotic-stage anthers from msh4 and mus81 mutants that mediate the Type I and Type II meiotic recombination pathways, respectively. We show that heat stress reduces the number of expressed genes regardless of genotype. In addition, msh4 mutants have a distinct gene expression pattern compared to mus81 and wild type controls. Interestingly, ASY1, which encodes a HORMA domain protein that is a component of meiotic chromosome axes, is up-regulated in wild type and mus81 but not in msh4. In addition, SDS the meiosis-specific cyclin-like gene, DMC1 the meiosis-specific recombinase, SYN1/REC8 the meiosis-specific cohesion complex component, and SWI1 which functions in meiotic sister chromatid cohesion are up-regulated in all three genotypes. We also characterize 51 novel, previously unannotated transcripts, and show that their promoter regions are associated with A-rich meiotic recombination hotspot motifs.
Conclusions: Our transcriptomic analysis of msh4 and mus81 mutants enhances our understanding of how the Type I and Type II meiotic CO pathway respond to environmental temperature stress and might provide a strategy to manipulate recombination levels in plants.
Keywords: ASY1; Heat stress; MSH4; MUS81; Meiotic recombination; RNA-Seq.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis.Proc Natl Acad Sci U S A. 2020 Jun 16;117(24):13647-13658. doi: 10.1073/pnas.1921055117. Epub 2020 Jun 4. Proc Natl Acad Sci U S A. 2020. PMID: 32499315 Free PMC article.
-
MCM8 is required for a pathway of meiotic double-strand break repair independent of DMC1 in Arabidopsis thaliana.PLoS Genet. 2013;9(1):e1003165. doi: 10.1371/journal.pgen.1003165. Epub 2013 Jan 3. PLoS Genet. 2013. PMID: 23300481 Free PMC article.
-
Arabidopsis PTD is required for type I crossover formation and affects recombination frequency in two different chromosomal regions.J Genet Genomics. 2014 Mar 20;41(3):165-75. doi: 10.1016/j.jgg.2014.02.001. Epub 2014 Feb 22. J Genet Genomics. 2014. PMID: 24656236
-
ASY1 coordinates early events in the plant meiotic recombination pathway.Cytogenet Genome Res. 2008;120(3-4):302-12. doi: 10.1159/000121079. Epub 2008 May 23. Cytogenet Genome Res. 2008. PMID: 18504359 Review.
-
The biochemistry of early meiotic recombination intermediates.Cell Cycle. 2018;17(23):2520-2530. doi: 10.1080/15384101.2018.1553355. Epub 2018 Dec 10. Cell Cycle. 2018. PMID: 30482074 Free PMC article. Review.
Cited by
-
The plant early recombinosome: a high security complex to break DNA during meiosis.Plant Reprod. 2024 Dec;37(4):421-440. doi: 10.1007/s00497-024-00509-7. Epub 2024 Sep 27. Plant Reprod. 2024. PMID: 39331138 Free PMC article. Review.
-
Meiotic chromosome organization and its role in recombination and cancer.Curr Top Dev Biol. 2023;151:91-126. doi: 10.1016/bs.ctdb.2022.04.008. Epub 2022 Jun 20. Curr Top Dev Biol. 2023. PMID: 36681479 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

