Bradyrhizobium agreste sp. nov., Bradyrhizobium glycinis sp. nov. and Bradyrhizobium diversitatis sp. nov., isolated from a biodiversity hotspot of the genus Glycine in Western Australia
- PMID: 33709900
- PMCID: PMC8375429
- DOI: 10.1099/ijsem.0.004742
Bradyrhizobium agreste sp. nov., Bradyrhizobium glycinis sp. nov. and Bradyrhizobium diversitatis sp. nov., isolated from a biodiversity hotspot of the genus Glycine in Western Australia
Abstract
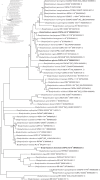
Strains of the genus Bradyrhizobium associated with agronomically important crops such as soybean (Glycine max) are increasingly studied; however, information about symbionts of wild Glycine species is scarce. Australia is a genetic centre of wild Glycine species and we performed a polyphasic analysis of three Bradyrhizobium strains-CNPSo 4010T, CNPSo 4016T, and CNPSo 4019T-trapped from Western Australian soils with Glycine clandestina, Glycine tabacina and Glycine max, respectively. The phylogenetic tree of the 16S rRNA gene clustered all strains into the Bradyrhizobium japonicum superclade; strains CNPSo 4010T and CNPSo 4016T had Bradyrhizobium yuanmingense CCBAU 10071T as the closest species, whereas strain CNPSo 4019T was closer to Bradyrhizobium liaoningense LMG 18230T. The multilocus sequence analysis (MLSA) with five housekeeping genes-dnaK, glnII, gyrB, recA and rpoB-confirmed the same clusters as the 16S rRNA phylogeny, but indicated low similarity to described species, with nucleotide identities ranging from 93.6 to 97.6% of similarity. Considering the genomes of the three strains, the average nucleotide identity and digital DNA-DNA hybridization values were lower than 94.97 and 59.80 %, respectively, with the closest species. In the nodC phylogeny, strains CNPSo 4010T and CNPSo 4019T grouped with Bradyrhizobium zhanjiangense and Bradyrhizobium ganzhouense, respectively, while strain CNPSo 4016T was positioned separately from the all symbiotic Bradyrhizobium species. Other genomic (BOX-PCR), phenotypic and symbiotic properties were evaluated and corroborated with the description of three new lineages of Bradyrhizobium. We propose the names of Bradyrhizobium agreste sp. nov. for CNPSo 4010T (=WSM 4802T=LMG 31645T) isolated from Glycine clandestina, Bradyrhizobium glycinis sp. nov. for CNPSo 4016T (=WSM 4801T=LMG 31649T) isolated from Glycine tabacina and Bradyrhizobium diversitatis sp. nov. for CNPSo 4019T (=WSM 4799T=LMG 31650T) isolated from G. max.
Keywords: ANI; Bradyrhizobium; Glycine; MLSA; dDDH; nodulation; wild soybean.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Bradyrhizobium cenepequi sp. nov., Bradyrhizobium semiaridum sp. nov., Bradyrhizobium hereditatis sp. nov. and Bradyrhizobium australafricanum sp. nov., symbionts of different leguminous plants of Western Australia and South Africa and definition of three novel symbiovars.Int J Syst Evol Microbiol. 2022 Jul;72(7). doi: 10.1099/ijsem.0.005446. Int J Syst Evol Microbiol. 2022. PMID: 35796350
-
Bradyrhizobium archetypum sp. nov., Bradyrhizobium australiense sp. nov. and Bradyrhizobium murdochi sp. nov., isolated from nodules of legumes indigenous to Western Australia.Int J Syst Evol Microbiol. 2020 Aug;70(8):4623-4636. doi: 10.1099/ijsem.0.004322. Int J Syst Evol Microbiol. 2020. PMID: 32667875
-
Bradyrhizobium niftali sp. nov., an effective nitrogen-fixing symbiont of partridge pea [Chamaecrista fasciculata (Michx.) Greene], a native caesalpinioid legume broadly distributed in the USA.Int J Syst Evol Microbiol. 2019 Nov;69(11):3448-3459. doi: 10.1099/ijsem.0.003640. Int J Syst Evol Microbiol. 2019. PMID: 31429819
-
Bradyrhizobium tropiciagri sp. nov. and Bradyrhizobium embrapense sp. nov., nitrogen-fixing symbionts of tropical forage legumes.Int J Syst Evol Microbiol. 2015 Dec;65(12):4424-4433. doi: 10.1099/ijsem.0.000592. Epub 2015 Sep 10. Int J Syst Evol Microbiol. 2015. PMID: 26362866
-
Bradyrhizobium frederickii sp. nov., a nitrogen-fixing lineage isolated from nodules of the caesalpinioid species Chamaecrista fasciculata and characterized by tolerance to high temperature in vitro.Int J Syst Evol Microbiol. 2019 Dec;69(12):3863-3877. doi: 10.1099/ijsem.0.003697. Int J Syst Evol Microbiol. 2019. PMID: 31486763
Cited by
-
New Insights into the Taxonomy of Bacteria in the Genomic Era and a Case Study with Rhizobia.Int J Microbiol. 2022 May 21;2022:4623713. doi: 10.1155/2022/4623713. eCollection 2022. Int J Microbiol. 2022. PMID: 35637770 Free PMC article. Review.
References
-
- De Bruijn FJ. Biological nitrogen fixation. In: Lugtenberg B, editor. Principles of Plant-Microbe Interactions: Microbes for Sustainable Agriculture. 1st ed. Switzerland: Springer International Publishing; 2015. pp. 1–448. editor.
-
- Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ. Bacterial associations with legumes. CRC Crit Rev Plant Sci. 2015;34:17–42.
-
- Ormeño-Orrillo E, Hungria M, Martinez-Romero E. Dinitrogen-fixing prokaryotes. In: Rosenberg E, DeLong E, Stackebrandt E, Lory S, Thompson F, editors. The Prokaryotes: Prokaryotic Physiology and Biochemistry. 4th ed. Berlin Heidelberg: Springer-Verlag; 2013. pp. 427–451.
LinkOut - more resources
Full Text Sources

