Adult astrocytes from reptiles are resistant to proinflammatory activation via sustaining Vav1 expression
- PMID: 33705794
- PMCID: PMC8065226
- DOI: 10.1016/j.jbc.2021.100527
Adult astrocytes from reptiles are resistant to proinflammatory activation via sustaining Vav1 expression
Abstract
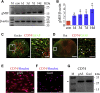
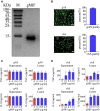
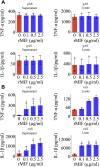
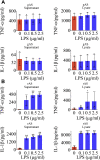
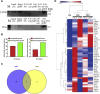
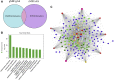
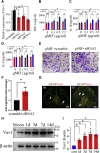
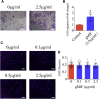
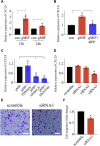
Adult mammalian astrocytes are sensitive to inflammatory stimuli in the context of neuropathology or mechanical injury, thereby affecting functional outcomes of the central nervous system (CNS). In contrast, glial cells residing in the spinal cord of regenerative vertebrates exhibit a weak astroglial reaction similar to those of mammals in embryonic stages. Macrophage migration inhibitory factor (MIF) participates in multiple neurological disorders by activation of glial and immune cells. However, the mechanism of astrocytes from regenerative species, such as gecko astrocytes (gAS), in resistance to MIF-mediated inflammation in the severed cords remains unclear. Here, we compared neural stem cell markers among gAS, as well as adult (rAS) and embryonic (eAS) rat astrocytes. We observed that gAS retained an immature phenotype resembling rat eAS. Proinflammatory activation of gAS with gecko (gMIF) or rat (rMIF) recombinant protein was unable to induce the production of inflammatory cytokines, despite its interaction with membrane CD74 receptor. Using cross-species screening of inflammation-related mediators from models of gMIF- and rMIF-induced gAS and rAS, we identified Vav1 as a key regulator in suppressing the inflammatory activation of gAS. The gAS with Vav1 deficiency displayed significantly restored sensitivity to inflammatory stimuli. Meanwhile, gMIF acts to promote the migration of gAS through regulation of CXCL8 following cord lesion. Taken together, our results suggest that Vav1 contributes to the regulation of astrocyte-mediated inflammation, which might be beneficial for the therapeutic development of neurological diseases.
Keywords: MIF; Vav1; astrocyte; cell migration; cytokine; immunosuppression; neuroinflammation; reptile; spinal cord.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors have declared that no competing interests exist.
Figures
Similar articles
-
Macrophage Migration Inhibitory Factor Promotes Expression of Matrix Metalloproteinases 1 and 3 in Spinal Cord Astrocytes following Gecko Tail Amputation.J Integr Neurosci. 2023 Feb 10;22(2):29. doi: 10.31083/j.jin2202029. J Integr Neurosci. 2023. PMID: 36992581
-
Macrophage migration inhibitory factor activates inflammatory responses of astrocytes through interaction with CD74 receptor.Oncotarget. 2017 Jan 10;8(2):2719-2730. doi: 10.18632/oncotarget.13739. Oncotarget. 2017. PMID: 27926507 Free PMC article.
-
Macrophage migration inhibitory factor facilitates production of CCL5 in astrocytes following rat spinal cord injury.J Neuroinflammation. 2018 Sep 4;15(1):253. doi: 10.1186/s12974-018-1297-z. J Neuroinflammation. 2018. PMID: 30180853 Free PMC article.
-
AKR1B1 Upregulation Contributes to Neuroinflammation and Astrocytes Proliferation by Regulating the Energy Metabolism in Rat Spinal Cord Injury.Neurochem Res. 2018 Aug;43(8):1491-1499. doi: 10.1007/s11064-018-2570-3. Epub 2018 Jun 12. Neurochem Res. 2018. PMID: 29948725 Review.
-
The Role of MIF on Eosinophil Biology and Eosinophilic Inflammation.Clin Rev Allergy Immunol. 2020 Feb;58(1):15-24. doi: 10.1007/s12016-019-08726-z. Clin Rev Allergy Immunol. 2020. PMID: 30680604 Review.
Cited by
-
Combination of single-cell and bulk RNA seq reveals the immune infiltration landscape and targeted therapeutic drugs in spinal cord injury.Front Immunol. 2023 Jan 19;14:1068359. doi: 10.3389/fimmu.2023.1068359. eCollection 2023. Front Immunol. 2023. PMID: 36742334 Free PMC article.
-
Reactive Astrocytes in Central Nervous System Injury: Subgroup and Potential Therapy.Front Cell Neurosci. 2021 Dec 23;15:792764. doi: 10.3389/fncel.2021.792764. eCollection 2021. Front Cell Neurosci. 2021. PMID: 35002629 Free PMC article. Review.
-
Self-Control of Inflammation during Tail Regeneration of Lizards.J Dev Biol. 2021 Nov 2;9(4):48. doi: 10.3390/jdb9040048. J Dev Biol. 2021. PMID: 34842738 Free PMC article. Review.
-
Evolution of astrocytes: From invertebrates to vertebrates.Front Cell Dev Biol. 2022 Aug 15;10:931311. doi: 10.3389/fcell.2022.931311. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36046339 Free PMC article. Review.
-
Astrocyte evolution and human specificity.Neural Regen Res. 2023 Jan;18(1):131-132. doi: 10.4103/1673-5374.340405. Neural Regen Res. 2023. PMID: 35799529 Free PMC article. No abstract available.
References
-
- Pekny M., Pekna M. Astrocyte reactivity and reactive astrogliosis: Costs and benefits. Physiol. Rev. 2014;94:1077–1098. - PubMed
-
- Colombo E., Farina C. Astrocytes: Key regulators of neuroinflammation. Trends Immunol. 2016;37:608–620. - PubMed
-
- Farina C., Aloisi F., Meinl E. Astrocytes are active players in cerebral innate immunity. Trends Immunol. 2007;28:138–145. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

