Persistent Human Papillomavirus Infection
- PMID: 33672465
- PMCID: PMC7923415
- DOI: 10.3390/v13020321
Persistent Human Papillomavirus Infection
Abstract
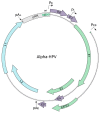
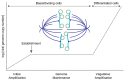
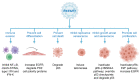
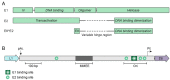
Persistent infection with oncogenic human papillomavirus (HPV) types is responsible for ~5% of human cancers. The HPV infectious cycle can sustain long-term infection in stratified epithelia because viral DNA is maintained as low copy number extrachromosomal plasmids in the dividing basal cells of a lesion, while progeny viral genomes are amplified to large numbers in differentiated superficial cells. The viral E1 and E2 proteins initiate viral DNA replication and maintain and partition viral genomes, in concert with the cellular replication machinery. Additionally, the E5, E6, and E7 proteins are required to evade host immune responses and to produce a cellular environment that supports viral DNA replication. An unfortunate consequence of the manipulation of cellular proliferation and differentiation is that cells become at high risk for carcinogenesis.
Keywords: HPV; cancer; epithelium; extrachromosomal replication; immune evasion; latency; papillomavirus; persistence; tethering.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript, or in the decision to publish the review.
Figures


Similar articles
-
Roles for E1-independent replication and E6-mediated p53 degradation during low-risk and high-risk human papillomavirus genome maintenance.PLoS Pathog. 2019 May 13;15(5):e1007755. doi: 10.1371/journal.ppat.1007755. eCollection 2019 May. PLoS Pathog. 2019. PMID: 31083694 Free PMC article.
-
Persistence of an Oncogenic Papillomavirus Genome Requires cis Elements from the Viral Transcriptional Enhancer.mBio. 2017 Nov 21;8(6):e01758-17. doi: 10.1128/mBio.01758-17. mBio. 2017. PMID: 29162712 Free PMC article.
-
FANCD2 Binds Human Papillomavirus Genomes and Associates with a Distinct Set of DNA Repair Proteins to Regulate Viral Replication.mBio. 2017 Feb 14;8(1):e02340-16. doi: 10.1128/mBio.02340-16. mBio. 2017. PMID: 28196964 Free PMC article.
-
Replication and partitioning of papillomavirus genomes.Adv Virus Res. 2008;72:155-205. doi: 10.1016/S0065-3527(08)00404-1. Adv Virus Res. 2008. PMID: 19081491 Free PMC article. Review.
-
Evasion of host immune defenses by human papillomavirus.Virus Res. 2017 Mar 2;231:21-33. doi: 10.1016/j.virusres.2016.11.023. Epub 2016 Nov 24. Virus Res. 2017. PMID: 27890631 Free PMC article. Review.
Cited by
-
p53-dependent R-loop formation and HPV pathogenesis.Proc Natl Acad Sci U S A. 2023 Aug 29;120(35):e2305907120. doi: 10.1073/pnas.2305907120. Epub 2023 Aug 23. Proc Natl Acad Sci U S A. 2023. PMID: 37611058 Free PMC article.
-
Functional roles of female sex hormones and their nuclear receptors in cervical cancer.Essays Biochem. 2021 Dec 17;65(6):941-950. doi: 10.1042/EBC20200175. Essays Biochem. 2021. PMID: 34156060 Free PMC article.
-
HPV-16 E7 Interacts with the Endocytic Machinery via the AP2 Adaptor μ2 Subunit.mBio. 2022 Dec 20;13(6):e0230222. doi: 10.1128/mbio.02302-22. Epub 2022 Oct 18. mBio. 2022. PMID: 36255238 Free PMC article.
-
Human Papillomavirus Carcinogenicity and the Need of New Perspectives: Thoughts from a Retrospective Analysis on Human Papillomavirus Outcomes Conducted at the Hospital University of Bari, Apulia, Italy, between 2011 and 2022.Diagnostics (Basel). 2024 May 6;14(9):968. doi: 10.3390/diagnostics14090968. Diagnostics (Basel). 2024. PMID: 38732382 Free PMC article.
-
Regulation of the Innate Immune Response during the Human Papillomavirus Life Cycle.Viruses. 2022 Aug 17;14(8):1797. doi: 10.3390/v14081797. Viruses. 2022. PMID: 36016419 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

