Participation of TFIIIB Subunit Brf1 in Transcription Regulation in the Human Pathogen Leishmania major
- PMID: 33669344
- PMCID: PMC7920299
- DOI: 10.3390/genes12020280
Participation of TFIIIB Subunit Brf1 in Transcription Regulation in the Human Pathogen Leishmania major
Abstract
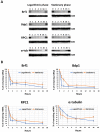
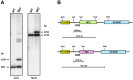
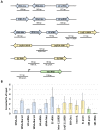
In yeast and higher eukaryotes, transcription factor TFIIIB is required for accurate initiation of transcription by RNA Polymerase III (Pol III), which synthesizes transfer RNAs (tRNAs), 5S ribosomal RNA (rRNA), and other essential RNA molecules. TFIIIB is composed of three subunits: B double prime 1 (Bdp1), TATA-binding protein (TBP), and TFIIB-related factor 1 (Brf1). Here, we report the molecular characterization of Brf1 in Leishmania major (LmBrf1), a parasitic protozoan that shows distinctive transcription characteristics, including the apparent absence of Pol III general transcription factors TFIIIA and TFIIIC. Although single-knockout parasites of LmBrf1 were obtained, attempts to generate LmBrf1-null mutants were unsuccessful, which suggests that LmBrf1 is essential in promastigotes of L. major. Notably, Northern blot analyses showed that the half-lives of the messenger RNAs (mRNAs) from LmBrf1 and other components of the Pol III transcription machinery (Bdp1 and Pol III subunit RPC1) are very similar (~40 min). Stabilization of these transcripts was observed in stationary-phase parasites. Chromatin immunoprecipitation (ChIP) experiments showed that LmBrf1 binds to tRNA, small nuclear RNA (snRNA), and 5S rRNA genes. Unexpectedly, the results also indicated that LmBrf1 associates to the promoter region of the 18S rRNA genes and to three Pol II-dependent regions here analyzed. Tandem affinity purification and mass spectrometry analyses allowed the identification of a putative TFIIIC subunit. Moreover, several proteins involved in transcription by all three RNA polymerases co-purified with the tagged version of LmBrf1.
Keywords: 5S rRNA; Leishmania; Pol III transcription; RNA polymerases; TFIIIB; tRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
TFIIIB Subunit Bdp1 Participates in RNA Polymerase III Transcription in the Protozoan Parasite Leishmania major.Biomed Res Int. 2019 Apr 1;2019:1425281. doi: 10.1155/2019/1425281. eCollection 2019. Biomed Res Int. 2019. PMID: 31058184 Free PMC article.
-
Structure-function analysis of the human TFIIB-related factor II protein reveals an essential role for the C-terminal domain in RNA polymerase III transcription.Mol Cell Biol. 2005 Nov;25(21):9406-18. doi: 10.1128/MCB.25.21.9406-9418.2005. Mol Cell Biol. 2005. PMID: 16227591 Free PMC article.
-
Differential expression of the TFIIIB subunits Brf1 and Brf2 in cancer cells.BMC Mol Biol. 2008 Aug 12;9:74. doi: 10.1186/1471-2199-9-74. BMC Mol Biol. 2008. PMID: 18700021 Free PMC article.
-
Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human.Nucleic Acids Res. 2001 Jul 1;29(13):2675-90. doi: 10.1093/nar/29.13.2675. Nucleic Acids Res. 2001. PMID: 11433012 Free PMC article. Review.
-
Regulation of tRNA synthesis by the general transcription factors of RNA polymerase III - TFIIIB and TFIIIC, and by the MAF1 protein.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):320-329. doi: 10.1016/j.bbagrm.2018.01.011. Epub 2018 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29378333 Review.
Cited by
-
Characterization of Tau95 led to the identification of a four-subunit TFIIIC complex in trypanosomatid parasites.Appl Microbiol Biotechnol. 2024 Dec;108(1):109. doi: 10.1007/s00253-023-12903-8. Epub 2024 Jan 10. Appl Microbiol Biotechnol. 2024. PMID: 38204130 Free PMC article.
-
Molecular Response of Meyerozyma guilliermondii to Patulin: Transcriptomic-Based Analysis.J Fungi (Basel). 2023 Apr 30;9(5):538. doi: 10.3390/jof9050538. J Fungi (Basel). 2023. PMID: 37233249 Free PMC article.
References
-
- Gunzl A., Vanhamme L., Myler P.J. Transcription in trypanosomes: A different means to the end. In: Barry J.D., McCulloch R., Mottram J.C., Acosta-Serrano A., editors. Trypanosomes: After the Genome. Horizon Bioscience; Wymonham, UK: 2007. pp. 177–208.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

