circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression
- PMID: 33664993
- PMCID: PMC7901030
- DOI: 10.1016/j.omtn.2020.12.013
circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression
Abstract
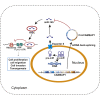
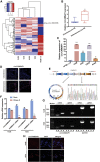
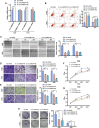
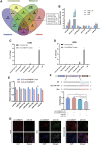
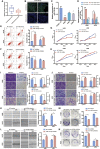
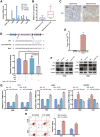
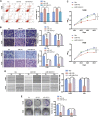
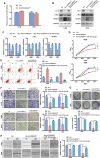
Osteosarcoma is the most common primary malignant bone tumor in adolescents. While chemotherapy combined with surgery can improve the prognosis of some patients, chemo-resistance is still a huge obstacle in osteosarcoma treatment. Accumulating evidence demonstrates that circular RNAs (circRNAs) are involved in cancer progression and metastasis, but their specific role in osteosarcoma remains mostly undescribed. In this study, we performed circRNA deep sequencing and identified 88 distinct circRNAs from a human osteosarcoma cell lines group (143B, HOS, SJSA, and U2OS) and the human osteoblast hFOB 1.19 (control). We found that circCAMSAP1, also named hsa_circ_0004338, is significantly upregulated in human osteosarcoma tissues and cell lines, and it is positively correlated with osteosarcoma development. Silencing of circCAMSAP1 effectively suppresses osteosarcoma cell growth, apoptosis, migration, and invasion. Furthermore, we validated that circCAMSAP1 functions in osteosarcoma tumorigenesis through a circCAMSAP1/miR-145-5p/friend leukemia virus integration 1 (FLI1) pathway. FLI1 promotes osteosarcoma tumorigenesis and miR-145-5p suppresses FLI translation. circCAMSAP1 directly sequesters miR-145-5p in the cytoplasm and inhibits its activity to suppress osteosarcoma tumorigenesis. Moreover, the regulatory role of circCAMSAP1 upregulation was examined and validated in rats. In summary, our findings provide evidence that circCAMSAP1 act as a "microRNA sponge" and suggest a new therapeutic target of human osteosarcoma.
© 2020.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
circCAMSAP1 Promotes Tumor Growth in Colorectal Cancer via the miR-328-5p/E2F1 Axis.Mol Ther. 2020 Mar 4;28(3):914-928. doi: 10.1016/j.ymthe.2019.12.008. Epub 2019 Dec 25. Mol Ther. 2020. PMID: 31951832 Free PMC article.
-
Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression.Mol Cancer. 2019 Apr 2;18(1):73. doi: 10.1186/s12943-019-1007-1. Mol Cancer. 2019. PMID: 30940151 Free PMC article.
-
Hsa_circ_0051079 functions as an oncogene by regulating miR-26a-5p/TGF-β1 in osteosarcoma.Cell Biosci. 2019 Nov 29;9:94. doi: 10.1186/s13578-019-0355-2. eCollection 2019. Cell Biosci. 2019. PMID: 31798828 Free PMC article.
-
Therapeutic Potential of Circular RNAs in Osteosarcoma.Front Oncol. 2020 Apr 15;10:370. doi: 10.3389/fonc.2020.00370. eCollection 2020. Front Oncol. 2020. PMID: 32351876 Free PMC article. Review.
-
MicroRNA signatures in osteosarcoma: diagnostic insights and therapeutic prospects.Mol Cell Biochem. 2024 Oct 17. doi: 10.1007/s11010-024-05135-5. Online ahead of print. Mol Cell Biochem. 2024. PMID: 39419925 Review.
Cited by
-
CircDOCK1 promotes the tumorigenesis and cisplatin resistance of osteogenic sarcoma via the miR-339-3p/IGF1R axis.Mol Cancer. 2021 Dec 7;20(1):161. doi: 10.1186/s12943-021-01453-0. Mol Cancer. 2021. PMID: 34876132 Free PMC article.
-
Alteration in the Expression of Circular Rnas and its association with the Development and Progression of Osteosarcoma, an Integrative Review with High Sensitivity Research.Asian Pac J Cancer Prev. 2024 Apr 1;25(4):1195-1203. doi: 10.31557/APJCP.2024.25.4.1195. Asian Pac J Cancer Prev. 2024. PMID: 38679978 Free PMC article. Review.
-
Hsa_circ_0001017 promotes cell proliferation, migration and invasion in osteosarcoma by sponging miR-145-5p.J Orthop Surg Res. 2022 Mar 28;17(1):184. doi: 10.1186/s13018-022-03062-z. J Orthop Surg Res. 2022. PMID: 35346268 Free PMC article.
-
CircNRIP1 Encapsulated by Bone Marrow Mesenchymal Stem Cell-Derived Extracellular Vesicles Aggravates Osteosarcoma by Modulating the miR-532-3p/AKT3/PI3K/AKT Axis.Front Oncol. 2021 Sep 29;11:658139. doi: 10.3389/fonc.2021.658139. eCollection 2021. Front Oncol. 2021. PMID: 34660257 Free PMC article.
-
Circular RNA CircPVT1 Inhibits 5-Fluorouracil Chemosensitivity by Regulating Ferroptosis Through MiR-30a-5p/FZD3 Axis in Esophageal Cancer Cells.Front Oncol. 2021 Dec 13;11:780938. doi: 10.3389/fonc.2021.780938. eCollection 2021. Front Oncol. 2021. PMID: 34966683 Free PMC article.
References
-
- Chou A.J., Geller D.S., Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr. Drugs. 2008;10:315–327. - PubMed
-
- Kaste S.C., Pratt C.B., Cain A.M., Jones-Wallace D.J., Rao B.N. Metastases detected at the time of diagnosis of primary pediatric extremity osteosarcoma at diagnosis: imaging features. Cancer. 1999;86:1602–1608. - PubMed
-
- Provisor A.J., Ettinger L.J., Nachman J.B., Krailo M.D., Makley J.T., Yunis E.J., Huvos A.G., Betcher D.L., Baum E.S., Kisker C.T., Miser J.S. Treatment of nonmetastatic osteosarcoma of the extremity with preoperative and postoperative chemotherapy: a report from the Children’s Cancer Group. J. Clin. Oncol. 1997;15:76–84. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

