How arginine derivatives alter the stability of lipid membranes: dissecting the roles of side chains, backbone and termini
- PMID: 33661339
- PMCID: PMC8071801
- DOI: 10.1007/s00249-021-01503-x
How arginine derivatives alter the stability of lipid membranes: dissecting the roles of side chains, backbone and termini
Abstract
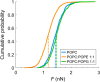
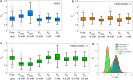
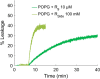
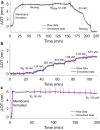
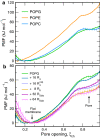
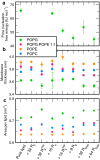
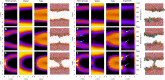
Arginine (R)-rich peptides constitute the most relevant class of cell-penetrating peptides and other membrane-active peptides that can translocate across the cell membrane or generate defects in lipid bilayers such as water-filled pores. The mode of action of R-rich peptides remains a topic of controversy, mainly because a quantitative and energetic understanding of arginine effects on membrane stability is lacking. Here, we explore the ability of several oligo-arginines R[Formula: see text] and of an arginine side chain mimic R[Formula: see text] to induce pore formation in lipid bilayers employing MD simulations, free-energy calculations, breakthrough force spectroscopy and leakage assays. Our experiments reveal that R[Formula: see text] but not R[Formula: see text] reduces the line tension of a membrane with anionic lipids. While R[Formula: see text] peptides form a layer on top of a partly negatively charged lipid bilayer, R[Formula: see text] leads to its disintegration. Complementary, our simulations show R[Formula: see text] causes membrane thinning and area per lipid increase beside lowering the pore nucleation free energy. Model polyarginine R[Formula: see text] similarly promoted pore formation in simulations, but without overall bilayer destabilization. We conclude that while the guanidine moiety is intrinsically membrane-disruptive, poly-arginines favor pore formation in negatively charged membranes via a different mechanism. Pore formation by R-rich peptides seems to be counteracted by lipids with PC headgroups. We found that long R[Formula: see text] and R[Formula: see text] but not short R[Formula: see text] reduce the free energy of nucleating a pore. In short R[Formula: see text], the substantial effect of the charged termini prevent their membrane activity, rationalizing why only longer [Formula: see text] are membrane-active.
Keywords: Arginine; Breakthrough force; CPP; MD-simulation.
Figures

Similar articles
-
The importance of membrane defects-lessons from simulations.Acc Chem Res. 2014 Aug 19;47(8):2244-51. doi: 10.1021/ar4002729. Epub 2014 Jun 3. Acc Chem Res. 2014. PMID: 24892900
-
Effect of lipid saturation on amyloid-beta peptide partitioning and aggregation in neuronal membranes: molecular dynamics simulations.Eur Biophys J. 2019 Dec;48(8):813-824. doi: 10.1007/s00249-019-01407-x. Epub 2019 Oct 26. Eur Biophys J. 2019. PMID: 31655893 Free PMC article.
-
Structural and Thermodynamic Insight into Spontaneous Membrane-Translocating Peptides Across Model PC/PG Lipid Bilayers.J Membr Biol. 2015 Jun;248(3):505-15. doi: 10.1007/s00232-014-9702-8. Epub 2014 Jul 10. J Membr Biol. 2015. PMID: 25008278
-
Arginine in membranes: the connection between molecular dynamics simulations and translocon-mediated insertion experiments.J Membr Biol. 2011 Jan;239(1-2):35-48. doi: 10.1007/s00232-010-9330-x. Epub 2010 Dec 3. J Membr Biol. 2011. PMID: 21127848 Free PMC article. Review.
-
A look at arginine in membranes.J Membr Biol. 2011 Jan;239(1-2):49-56. doi: 10.1007/s00232-010-9323-9. Epub 2010 Nov 25. J Membr Biol. 2011. PMID: 21107547 Free PMC article. Review.
Cited by
-
Molecular dynamics study of the internalization of cell-penetrating peptides containing unnatural amino acids across membranes.Nanoscale Adv. 2021 Nov 10;4(2):397-407. doi: 10.1039/d1na00674f. eCollection 2022 Jan 18. Nanoscale Adv. 2021. PMID: 36132688 Free PMC article.
-
Peptide-based pore formation and cell membrane deformation: European Biophysics Journal Prizes at EBSA 2023.Eur Biophys J. 2023 Nov;52(8):619-623. doi: 10.1007/s00249-023-01691-8. Eur Biophys J. 2023. PMID: 37994943
-
Implicit model to capture electrostatic features of membrane environment.PLoS Comput Biol. 2024 Jan 22;20(1):e1011296. doi: 10.1371/journal.pcbi.1011296. eCollection 2024 Jan. PLoS Comput Biol. 2024. PMID: 38252688 Free PMC article.
-
Adsorption/Desorption of Cationic-Hydrophobic Peptides on Zwitterionic Lipid Bilayer Is Associated with the Possibility of Proton Transfer.Antibiotics (Basel). 2023 Jul 21;12(7):1216. doi: 10.3390/antibiotics12071216. Antibiotics (Basel). 2023. PMID: 37508312 Free PMC article.
-
Increasing Angiogenesis Factors in Hypoxic Diabetic Wound Conditions by siRNA Delivery: Additive Effect of LbL-Gold Nanocarriers and Desloratadine-Induced Lysosomal Escape.Int J Mol Sci. 2021 Aug 26;22(17):9216. doi: 10.3390/ijms22179216. Int J Mol Sci. 2021. PMID: 34502144 Free PMC article.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25.
-
- Allolio C, Magarkar A, Jurkiewicz P, Baxová K, Javanainen M, Mason PE, Šachl R, Cebecauer M, Hof M, Horinek D, Heinz V, Rachel R, Ziegler CM, Schröfel A, Jungwirth P. Arginine-rich cell-penetrating peptides induce membrane multilamellarity and subsequently enter via formation of a fusion pore. Proc Natl Acad Sci. 2018;115(47):11923. - PMC - PubMed
-
- Alvaro IH, John MT, Om P. Membrane interacting peptides: a review. Curr Protein Pept Sci. 2016;17(8):827–841. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

