Spatial decoding of endosomal cAMP signals by a metastable cytoplasmic PKA network
- PMID: 33649598
- PMCID: PMC8084946
- DOI: 10.1038/s41589-021-00747-0
Spatial decoding of endosomal cAMP signals by a metastable cytoplasmic PKA network
Abstract
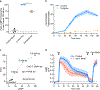
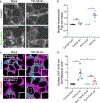
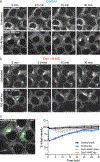
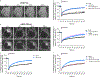
G-protein-coupled receptor-regulated cAMP production from endosomes can specify signaling to the nucleus by moving the source of cAMP without changing its overall amount. How this is possible remains unknown because cAMP gradients dissipate over the nanoscale, whereas endosomes typically localize micrometers from the nucleus. We show that the key location-dependent step for endosome-encoded transcriptional control is nuclear entry of cAMP-dependent protein kinase (PKA) catalytic subunits. These are sourced from punctate accumulations of PKA holoenzyme that are densely distributed in the cytoplasm and titrated by global cAMP into a discrete metastable state, in which catalytic subunits are bound but dynamically exchange. Mobile endosomes containing activated receptors collide with the metastable PKA puncta and pause in close contact. We propose that these properties enable cytoplasmic PKA to act collectively like a semiconductor, converting nanoscale cAMP gradients generated from endosomes into microscale elevations of free catalytic subunits to direct downstream signaling.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing interests.
Figures


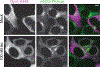
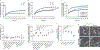
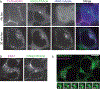


Similar articles
-
Novel, isotype-specific sensors for protein kinase A subunit interaction based on bioluminescence resonance energy transfer (BRET).Cell Signal. 2006 Oct;18(10):1616-25. doi: 10.1016/j.cellsig.2006.01.013. Epub 2006 Mar 9. Cell Signal. 2006. PMID: 16524697
-
Regulation of nuclear PKA revealed by spatiotemporal manipulation of cyclic AMP.Nat Chem Biol. 2012 Feb 26;8(4):375-82. doi: 10.1038/nchembio.799. Nat Chem Biol. 2012. PMID: 22366721 Free PMC article.
-
The differential response of protein kinase A to cyclic AMP in discrete brain areas correlates with the abundance of regulatory subunit II.J Neurochem. 1996 Apr;66(4):1752-61. doi: 10.1046/j.1471-4159.1996.66041752.x. J Neurochem. 1996. PMID: 8627334
-
Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design.Biochim Biophys Acta. 2008 Jan;1784(1):16-26. doi: 10.1016/j.bbapap.2007.10.002. Epub 2007 Oct 12. Biochim Biophys Acta. 2008. PMID: 17996741 Free PMC article. Review.
-
Specificity in the cAMP/PKA signaling pathway. Differential expression,regulation, and subcellular localization of subunits of PKA.Front Biosci. 2000 Aug 1;5:D678-93. doi: 10.2741/skalhegg. Front Biosci. 2000. PMID: 10922298 Review.
Cited by
-
An engineered trafficking biosensor reveals a role for DNAJC13 in DOR downregulation.Nat Chem Biol. 2024 Sep 2. doi: 10.1038/s41589-024-01705-2. Online ahead of print. Nat Chem Biol. 2024. PMID: 39223388
-
"Radical" differences between two FLIM microscopes affect interpretation of cell signaling dynamics.iScience. 2024 Jun 13;27(7):110268. doi: 10.1016/j.isci.2024.110268. eCollection 2024 Jul 19. iScience. 2024. PMID: 39036041 Free PMC article.
-
Non-canonical β-adrenergic activation of ERK at endosomes.Nature. 2022 Nov;611(7934):173-179. doi: 10.1038/s41586-022-05343-3. Epub 2022 Oct 26. Nature. 2022. PMID: 36289326 Free PMC article.
-
Spatial organization of adenylyl cyclase and its impact on dopamine signaling in neurons.Nat Commun. 2024 Sep 27;15(1):8297. doi: 10.1038/s41467-024-52575-0. Nat Commun. 2024. PMID: 39333071 Free PMC article.
-
Location bias contributes to functionally selective responses of biased CXCR3 agonists.Nat Commun. 2022 Oct 4;13(1):5846. doi: 10.1038/s41467-022-33569-2. Nat Commun. 2022. PMID: 36195635 Free PMC article.
References
-
- Calebiro D & Godbole A Internalization of G-protein-coupled receptors: Implication in receptor function, physiology and diseases. Best Pract. Res. Clin. Endocrinol. Metab 32, 83–91 (2018). - PubMed
METHODS REFERENCES
-
- Patriarchi T et al. Imaging neuromodulators with high spatiotemporal resolution using genetically encoded indicators. Nat. Protoc 14, 3471–3505 (2019). - PubMed
-
- Niwa H, Yamamura K & Miyazaki J Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

