CD8+ T-Cell Memory: The Why, the When, and the How
- PMID: 33648987
- PMCID: PMC8091951
- DOI: 10.1101/cshperspect.a038661
CD8+ T-Cell Memory: The Why, the When, and the How
Abstract
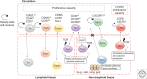
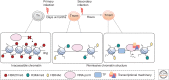
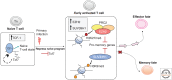
The generation of effective adaptive T-cell memory is a cardinal feature of the adaptive immune system. The establishment of protective T-cell immunity requires the differentiation of CD8+ T cells from a naive state to one where pathogen-specific memory CD8+ T cells are capable of responding to a secondary infection more rapidly and robustly without the need for further differentiation. The study of factors that determine the fate of activated CD8+ T cells into either effector or memory subsets has a long history. The advent of new technologies is now providing new insights into how epigenetic regulation not only impacts acquisition and maintenance of effector function, but also the maintenance of the quiescent yet primed memory state. There is growing appreciation that rather than distinct subsets, memory T-cell populations may reflect different points on a spectrum between the starting naive T-cell population and a terminally differentiated effector CD8+ T-cell population. Interestingly, there is growing evidence that the molecular mechanisms that underpin the rapid effector function of memory T cells are also observed in innate immune cells such as macrophages and natural killer (NK) cells. This raises an interesting hypothesis that the memory/effector T-cell state represents a default innate-like response to antigen recognition, and that it is the naive state that is the defining feature of adaptive immunity. These issues are discussed.
Copyright © 2021 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
Similar articles
-
Running to Stand Still: Naive CD8+ T Cells Actively Maintain a Program of Quiescence.Int J Mol Sci. 2020 Dec 21;21(24):9773. doi: 10.3390/ijms21249773. Int J Mol Sci. 2020. PMID: 33371448 Free PMC article. Review.
-
Motility Matters: How CD8+ T-Cell Trafficking Influences Effector and Memory Cell Differentiation.Cold Spring Harb Perspect Biol. 2021 Aug 2;13(8):a038075. doi: 10.1101/cshperspect.a038075. Cold Spring Harb Perspect Biol. 2021. PMID: 34001529 Free PMC article. Review.
-
The major CD8 T cell effector memory subset in the normal and Chlamydia trachomatis-infected human endocervix is low in perforin.BMC Immunol. 2012 Dec 7;13:66. doi: 10.1186/1471-2172-13-66. BMC Immunol. 2012. PMID: 23216954 Free PMC article.
-
Developmental Origin Governs CD8+ T Cell Fate Decisions during Infection.Cell. 2018 Jun 28;174(1):117-130.e14. doi: 10.1016/j.cell.2018.05.029. Epub 2018 Jun 14. Cell. 2018. PMID: 29909981
-
Memory CD8+ T cell differentiation.Ann N Y Acad Sci. 2010 Jan;1183:251-66. doi: 10.1111/j.1749-6632.2009.05126.x. Ann N Y Acad Sci. 2010. PMID: 20146720 Free PMC article. Review.
Cited by
-
Peripheral blood T-cell subset and its clinical significance in lupus nephritis patients.Lupus Sci Med. 2022 Aug;9(1):e000717. doi: 10.1136/lupus-2022-000717. Lupus Sci Med. 2022. PMID: 35973743 Free PMC article.
-
The contributions of T cell-mediated immunity to protection from vaccine-preventable diseases: A primer.Hum Vaccin Immunother. 2024 Dec 31;20(1):2395679. doi: 10.1080/21645515.2024.2395679. Epub 2024 Aug 29. Hum Vaccin Immunother. 2024. PMID: 39205626 Free PMC article. Review.
-
Vaccine development: obligate intracellular bacteria new tools, old pathogens: the current state of vaccines against obligate intracellular bacteria.Front Cell Infect Microbiol. 2024 Mar 19;14:1282183. doi: 10.3389/fcimb.2024.1282183. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38567021 Free PMC article. Review.
-
Exploration and validation of a combined Hypoxia and m6A/m5C/m1A regulated gene signature for prognosis prediction of liver cancer.BMC Genomics. 2023 Dec 14;24(1):776. doi: 10.1186/s12864-023-09876-3. BMC Genomics. 2023. PMID: 38097948 Free PMC article.
-
Causality between immunocytes and polymyositis: A Mendelian randomization analysis.Medicine (Baltimore). 2024 Oct 25;103(43):e40254. doi: 10.1097/MD.0000000000040254. Medicine (Baltimore). 2024. PMID: 39470507 Free PMC article.
References
-
- Anderson RJ, Li J, Kedzierski L, Compton BJ, Hayman CM, Osmond TL, Tang CW, Farrand KJ, Koay HF, Almeida C, et al. 2017. Augmenting influenza-specific T cell memory generation with a natural killer T cell-dependent glycolipid-peptide vaccine. ACS Chem Biol 12: 2898–2905. 10.1021/acschembio.7b00845 - DOI - PubMed
-
- Araki Y, Wang Z, Zang C, Wood WH, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, et al. 2009. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30: 912–925. 10.1016/j.immuni.2009.05.006 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
