A Mechanism Leading to Changes in Copy Number Variations Affected by Transcriptional Level Might Be Involved in Evolution, Embryonic Development, Senescence, and Oncogenesis Mediated by Retrotransposons
- PMID: 33644055
- PMCID: PMC7905054
- DOI: 10.3389/fcell.2021.618113
A Mechanism Leading to Changes in Copy Number Variations Affected by Transcriptional Level Might Be Involved in Evolution, Embryonic Development, Senescence, and Oncogenesis Mediated by Retrotransposons
Abstract
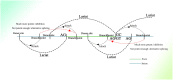
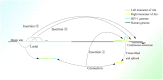
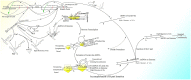
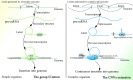
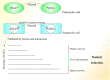
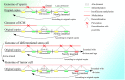
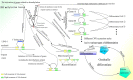
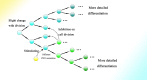
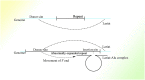
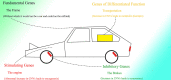
In recent years, more and more evidence has emerged showing that changes in copy number variations (CNVs) correlated with the transcriptional level can be found during evolution, embryonic development, and oncogenesis. However, the underlying mechanisms remain largely unknown. The success of the induced pluripotent stem cell suggests that genome changes could bring about transformations in protein expression and cell status; conversely, genome alterations generated during embryonic development and senescence might also be the result of genome changes. With rapid developments in science and technology, evidence of changes in the genome affected by transcriptional level has gradually been revealed, and a rational and concrete explanation is needed. Given the preference of the HIV-1 genome to insert into transposons of genes with high transcriptional levels, we propose a mechanism based on retrotransposons facilitated by specific pre-mRNA splicing style and homologous recombination (HR) to explain changes in CNVs in the genome. This mechanism is similar to that of the group II intron that originated much earlier. Under this proposed mechanism, CNVs on genome are dynamically and spontaneously extended in a manner that is positively correlated with transcriptional level or contract as the cell divides during evolution, embryonic development, senescence, and oncogenesis, propelling alterations in them. Besides, this mechanism explains several critical puzzles in these processes. From evidence collected to date, it can be deduced that the message contained in genome is not just three-dimensional but will become four-dimensional, carrying more genetic information.
Keywords: copy number variation; embryonic development; evolution; homologous recombination; oncogenesis; retrotransposons; senescence; transcription.
Copyright © 2021 Sui and Peng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Retrotransposons in pluripotent stem cells.Cell Regen. 2020 Jun 2;9(1):4. doi: 10.1186/s13619-020-00046-4. Cell Regen. 2020. PMID: 32588192 Free PMC article. Review.
-
Rapid Increase in frequency of gene copy-number variants during experimental evolution in Caenorhabditis elegans.BMC Genomics. 2015 Dec 9;16:1044. doi: 10.1186/s12864-015-2253-2. BMC Genomics. 2015. PMID: 26645535 Free PMC article.
-
Copy number alterations and copy number variation in cancer: close encounters of the bad kind.Cytogenet Genome Res. 2008;123(1-4):176-82. doi: 10.1159/000184706. Epub 2009 Mar 11. Cytogenet Genome Res. 2008. PMID: 19287153 Review.
-
Somatic mosaicism for copy-neutral loss of heterozygosity and DNA copy number variations in the human genome.BMC Genomics. 2015 Sep 16;16(1):703. doi: 10.1186/s12864-015-1916-3. BMC Genomics. 2015. PMID: 26376747 Free PMC article.
-
Interactions among genomic structure, function, and evolution revealed by comprehensive analysis of the Arabidopsis thaliana genome.Genomics. 2006 Oct;88(4):394-406. doi: 10.1016/j.ygeno.2006.05.003. Epub 2006 Jun 27. Genomics. 2006. PMID: 16806804
Cited by
-
Ovarian Cancer Patient-Derived Organoids Used as a Model for Replicating Genetic Characteristics and Testing Drug Responsiveness: A Preliminary Study.Cell Transplant. 2024 Jan-Dec;33:9636897241281869. doi: 10.1177/09636897241281869. Cell Transplant. 2024. PMID: 39323050 Free PMC article.
-
Duplication Versus Deletion Through the Lens of 15q13.3: Clinical and Research Implications of Studying Copy Number Variants Associated with Neuropsychiatric Disorders in Induced Pluripotent Stem Cell-Derived Neurons.Stem Cell Rev Rep. 2023 Apr;19(3):639-650. doi: 10.1007/s12015-022-10475-0. Epub 2022 Nov 12. Stem Cell Rev Rep. 2023. PMID: 36370261 Free PMC article. Review.
References
-
- Afridi A. K., Alam K. (2004). Prevalence and etiology of obesity–an overview. Pak. J. Nutr. 3, 14–25. 10.3923/pjn.2004.14.25 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

